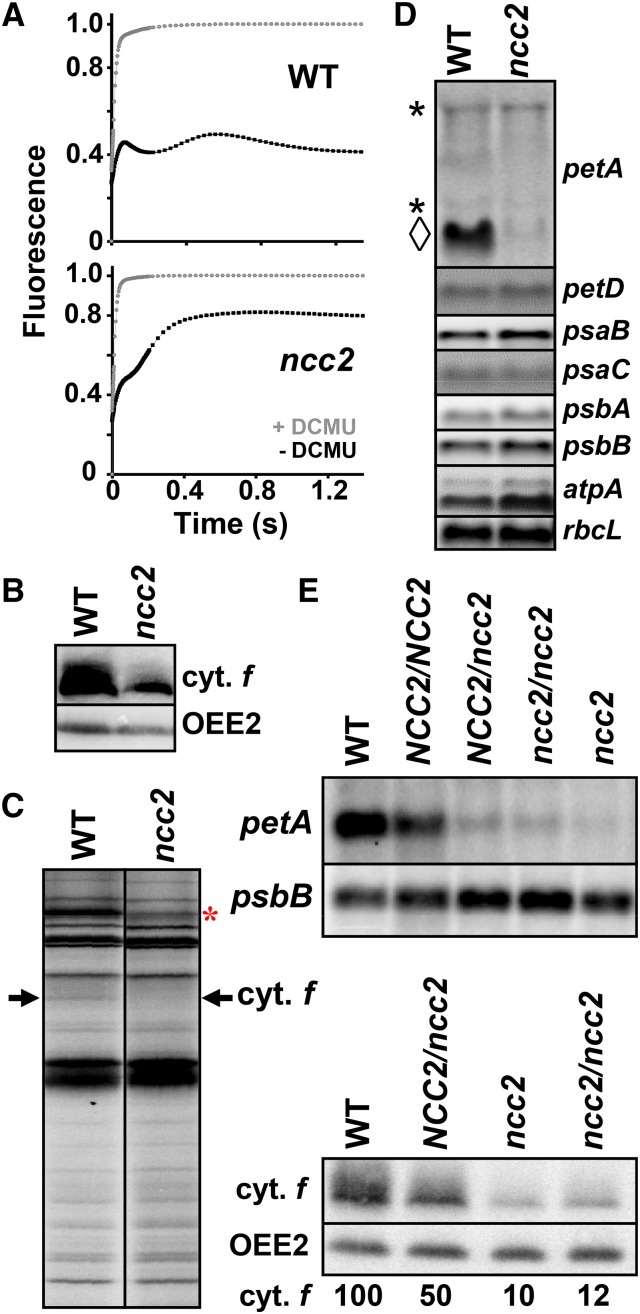
Figure 2.
The Dominant ncc2 Mutation Confers a b6f Leaky Phenotype Due to Reduced Accumulation of the petA mRNA.
(A) Fluorescence induction kinetics of dark-adapted ncc2 and wild-type cells. Black and gray curves show the kinetics recorded in the absence and in the presence, respectively, of DCMU (5 μM), which blocks electron transfer downstream of PSII. Maximal fluorescence levels in the presence of DCMU were normalized to 1.
(B) Cytochrome f accumulation in wild-type and ncc2 strains. OEE2 provides a loading control.
(C) Translation of chloroplast genes determined by 5 min 14C-acetate (5 μCi⋅mL−1) pulse-labeling experiments performed in the presence of cycloheximide (10 μg⋅mL−1) to block cytosolic translation. The arrow indicates the position of cytochrome f. The origin of the reduced rate of synthesis of Rubisco LS (red asterisk) in the ncc2 mutant, despite the unchanged accumulation of the rbcL transcript (D), is unknown.
(D) Accumulation of representative transcripts for chloroplast photosynthesis genes in wild-type and ncc2 strains, assessed by RNA gel blots using the probes indicated on the right of the panel. For the petA gene, the diamond indicates the mature transcript, while asterisks point to precursor RNA species.
(E) Accumulation of petA mRNA and cytochrome f, determined as above, in diploid strains either homozygous or heterozygous for the ncc2 mutation. Wild-type and ncc2 strains are shown for comparison. psbB mRNA and OEE2 provide loading controls in RNA and immunoblots, respectively.