Figure 1.
The Phosphatase Activity of Recombinant At-PPH1-SD Protein.
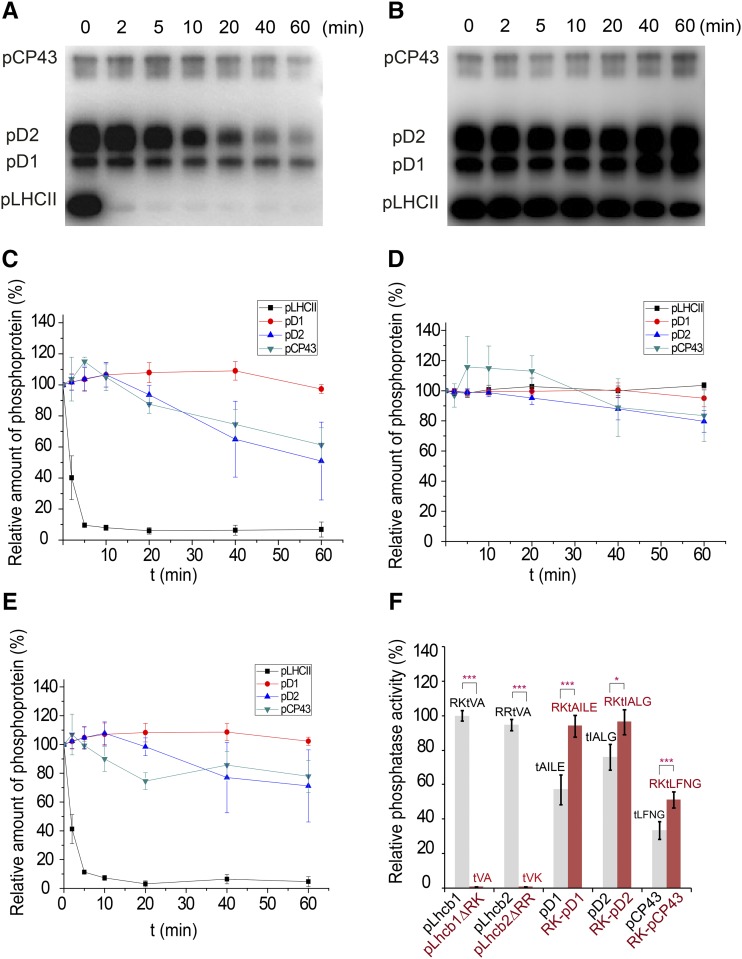
(A) Dephosphorylation of the thylakoid membrane proteins catalyzed by the purified PPH1-SD protein. The phosphoproteins were prepared from the pph1-2 plant and probed through the protein gel blot using the antiphosphothreonine antibody.
(B) A parallel control experiments were performed without adding the PPH1-SD protein in the reactions. The data show the background activities of endogenous thylakoid phosphatases other than TAP38/PPH1.
(C) The dephosphorylation kinetic curves of pLHCII, pD1, pD2, and pCP43 corresponding to the protein gel blot shown in (A). The intensities of the protein bands at different time points were scaled to those at the beginning of reactions. The relative amount of phosphoproteins = 100% when t = 0 min.
(D) The dephosphorylation kinetic curves corresponding to the control protein gel blot shown in (B).
(E) Processed kinetic curves representing the dephosphorylation activities catalyzed by exogenous PPH1-SD alone. The curves were plotted with the band intensity data subtracting the effects of endogenous phosphatases. The relative amount of phosphoproteins = 100% − ΔIPPH1. ΔIPPH1 represents the percentage of band intensity decrease caused by the activity of PPH1-SD. ΔIPPH1 = [I0(A) − It(A)]/I0(A) − [I0(B) − It(B)]/I0(B). I0(A) and I0(B) represents the 0-min band intensities extracted from A and B panels, respectively. It(A) and It(B) represents the t-min band intensities extracted from (A) and (B), respectively. The error bars in (C) to (E) indicate the standard errors of the mean values of three independent repeats (n = 3).
(F) The relative phosphatase activities of PPH1-SD toward different synthetic phosphorylated peptides. The activity of PPH1-SD was measured through the phosphatase assay system monitoring the release of free phosphate during the dephosphorylation reaction. Its activity toward pLhcb1 phosphopeptide (RKtVA) was normalized as 100% and the rest data were proportionally scaled. The sequences of the phosphopeptides (pLhcb1, pLhcb1ΔRK, pLhcb2, pLhcb2ΔRR, pD1, RK-pD1, pD2, RK-pD2, pCP43, and RK-pCP43) used for the assay are labeled on top of the columns. The error bars denote the standard errors of the mean values (n = 9 except for pLhcb1ΔRK and pLhcb2ΔRR, whose n = 6; *P < 0.05 and ***P < 0.001 by t test).