Fig. 3.
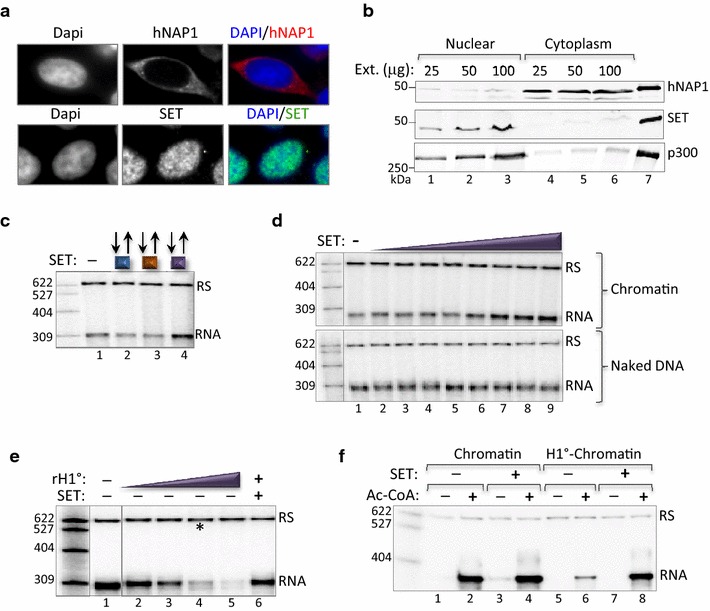
SET/Taf1β (SET), opposes the repressive function of H1 to enhance activated transcription in vitro. a Immunofluorescence labeling of hNAP1 (red, upper panel) and SET (green, lower panel) reveals distinct subcellular localization of the histone chaperones in HEK-293T cells. DAPI labeling of DNA is shown in blue. For single channel staining, black and white gave better resolution. For dual channel staining, color images were used to distinguish between the two antibodies. b Western blot analysis of hNAP1 and SET in nuclear and cytoplasmic extracts prepared from CEM T-cells. Immunostaining of nuclear p300 was performed to monitor efficiency of fractionation. Recombinant, purified hNAP1, SET, and p300 were analyzed in parallel as a positive control (lane 7). Recombinant hNAP1 migrates slightly slower due to the His6 purification tag. The position of the molecular weight size markers (kDa) is shown. c Like hNAP1, SET enhances activates transcription during the PIC assembly step. The in vitro transcription reaction was performed exactly as described for hNAP1 in Fig. 1e (lanes 9–12), in the presence of activators and SET (0.67 µM). d Dose-dependent enhancement of transcription by SET requires chromatin. In vitro transcription assay performed with increasing amounts of SET (0.03, 0.06, 0.12, 0.2, 0.27, 0.33 and 0.67 µM) added during PIC assembly produced a dose-dependent enhancement of transcription from chromatin templates (upper panel), but not from naked DNA (lower panel). The experiment was performed in the presence of activators and Ac-CoA. e SET reverses rH10-induced transcriptional repression. The in vitro transcription experiment was performed as described in Fig. 2a, with increasing amounts of rH10 (0.015, 0.03, 0.06, and 0.12 μM) (lanes 2–5), or a constant amount of rH10 (0.06 μM*) and SET (0.34 μM) (lane 6), in the presence of activators and Ac-CoA. f SET reverses transcriptional repression induced by H10-chromatin. In vitro transcription reactions were performed on the 900 bp HTLV-1/G-less cassette fragment, exactly as described for Fig. 2d, using SET (0.67 µM) in place of hNAP1. Reactions were performed in the presence of activators, and the absence or presence of SET and/or Ac-CoA, as indicated
