Abstract
Historically, genome-wide and molecular characterization of the genus Listeria has concentrated on the important human pathogen Listeria monocytogenes and a small number of closely related species, together termed Listeria sensu strictu. More recently, a number of genome sequences for more basal, and nonpathogenic, members of the Listeria genus have become available, facilitating a wider perspective on the evolution of pathogenicity and genome level evolutionary dynamics within the entire genus (termed Listeria sensu lato). Here, we have sequenced the genomes of additional Listeria fleischmannii and Listeria newyorkensis isolates and explored the dynamics of genome evolution in Listeria sensu lato. Our analyses suggest that acquisition of genetic material through gene duplication and divergence as well as through lateral gene transfer (mostly from outside Listeria) is widespread throughout the genus. Novel genetic material is apparently subject to rapid turnover. Multiple lines of evidence point to significant differences in evolutionary dynamics between the most basal Listeria subclade and all other congeners, including both sensu strictu and other sensu lato isolates. Strikingly, these differences are likely attributable to stochastic, population-level processes and contribute to observed variation in genome size across the genus. Notably, our analyses indicate that the common ancestor of Listeria sensu lato lacked flagella, which were acquired by lateral gene transfer by a common ancestor of Listeria grayi and Listeria sensu strictu, whereas a recently functionally characterized pathogenicity island, responsible for the capacity to produce cobalamin and utilize ethanolamine/propane-2-diol, was acquired in an ancestor of Listeria sensu strictu.
Keywords: Listeria, genome sequencing, comparative genomics, lateral gene transfer, cobalamin, ethanolamine metabolism, propane-2-diol metabolism, flagella
Introduction
Historically, genome-wide and molecular characterization of the genus Listeria has concentrated on the important human pathogen Listeria monocytogenes (for which the genomes of around 100 isolates have been sequenced) and a relatively restricted number of closely related species including Listeria marthii, Listeria innocua, Listeria welshimeri, Listeria seeligeri, and Listeria ivanovii. Together, these six species, all of which are capable of growth within higher vertebrate hosts, are known as Listeria sensu strictu, and various studies have focused on the identification and characterization of virulence factors that determine the differential capacity of many L. monocytogenes and L. ivanovii isolates to cause disease in mammals. Indeed, pathogenicity correlates well with the presence of intact virulence genes that facilitate direct interaction between bacteria and host cells—promoting colonization, entry into and death of host cells (Mishra et al. 2011). In particular, the virulence gene cluster (LIPI-1) promotes cytosolic replication as well as intra- and intercellular movement (Portnoy et al. 1992). A second cluster required for virulence contains an operon of two genes (inlA/B) that encode internalins necessary for the attachment to and invasion of nonphagocytic host cells (Hamon et al. 2006). Additionally L. ivanovii harbors a specific genetic island with virulence factors called LIPI-2, comprising of multiple internalins and the smcL sphingomyelinase hemolysis gene (Dominguez-Bernal et al. 2006). Phylogenomic analyses of Listeria sensu strictu suggest that nonpathogenic species evolved from a pathogenic ancestor through differential loss of pathogenicity factors (Schmid et al. 2005).
Besides the Listeria sensu strictu group, other, more divergent and presumably basal nonpathogenic species have long been known and more recently, many of these, as well as newly discovered congeners and representatives of the genus Brochothrix (Weller et al. 2014)—the presumed sister group to Listeria—have been subjected to whole-genome sequencing and phylogenetic analysis of concatenated core gene sets. Listeria grayi susbsp. grayi (DSM 20601, GenBank accession number ACCR00000000) and L. grayi subsp. murrayi (ATCC25401), isolated from vegetation (Welshimer and Meredith 1971) and sequenced by den Bakker et al. (2014), represent the sister taxon to the Listeria sensu strictu group. Listeria fleischmannii (susbsp. fleischmannii [from cheese] [Bertsch et al. 2012], subsp. coloradonensis [from agricultural environments] [den Bakker et al. 2013]) and L. fleischmannii FSL S10-1203 (den Bakker et al. 2014), along with Listeria floridensis and Listeria aquatica (both from running water) (den Bakker et al. 2014), constitute the sister clade to Listeria sensu strictu and L. grayi (Weller et al. 2014). Listeria rocourtiae (from lettuce leaves) (Leclercq et al. 2009; den Bakker et al. 2014), Listeria cornellensis, Listeria riparia and Listeria grandensis (all from water/agricultural environments) (den Bakker et al. 2014), Listeria weihenstephanensis (from aquatic vegetation) (Lang Halter et al. 2013; den Bakker et al. 2014), Listeria booriae (from seafood) (Weller et al. 2014), and Listeria newyorkensis (from a milk processing plant) (Weller et al. 2014) constitute the basal clade in the genus (Weller et al. 2014). Representatives of the L. fleischmannii and L. rocourtiae clades have been reported to be nonmotile (den Bakker et al. 2014). Their possible role as genetic reservoirs of genes that might be transferred to sensu strictu species remains to be investigated.
In this study, we have sequenced the genomes of additional L. fleischmannii and L. newyorkensis isolates. Probabilistic reconstruction of patterns of gene gain and loss, as well as comparative and phylogenomic surveys of all available basal Listeria genomes, indicates a nonpathogenic, nonmotile ancestor of extant Listeria species. Genome evolution is characterized by gene gain by lateral transfer (predominantly from outside the genus) and gene duplication—as well as by gene loss. Throughout Listeria sensu lato, genome size and dN/dS ratio show significant negative correlation, suggesting that rates of genetic drift underlie at least some of the observed variability in evolutionary dynamics. In particular, the larger genomes of the most basal subclade of Listeria are associated with an increased rate of gene gain with respect to other Listeria. Concentrated episodes of gene gain are inferred to have occurred along several major internal branches of the Listeria phylogeny, in particular, the entire flagella apparatus was acquired by a common ancestor of L. grayi and the Listeria sensu strictu clade, possibly through lateral gene transfer (LGT) from a Bacillus cereus-like organism. Furthermore, a recently characterized pathogenicity island encoding genes involved in ethanolamine/propane-2-diol/cobalamin metabolism was acquired and assembled, potentially from several donors, by a common ancestor of Listeria sensu strictu.
Materials and Methods
Whole-Genome Sequencing
Genomic DNA was extracted from the Listeria-like isolates using the QIAmp DNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. An indexed genomic library for each Listeria-like isolate was prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA), and a 2 × 250 paired-end sequencing run was performed on Illumina MiSeq platform.
Sequence Data Preprocessing
A custom script implementing strict quality filters based on the provided base call quality scores was used to trim sequences reads prior to assembly. Reads were iteratively trimmed from the 3′-end until all of the following conditions were satisfied:
The median quality score (Qscore) of upstream bases was >25.
Less than three bases with Qscore ≤10 and less than five bases with Qscore ≤20 were present in the upstream sequence.
The cumulative error probability in the upstream region was below 0.01.
Genome Assembly and Annotation
The velvet (Zerbino and Birney 2008) program was used for genome assembly. Optimal parameters for K-mers length and coverage were established manually, after careful inspection of the K-mer frequency graph for different values of K (21–81), as described in the manual. Scaffolding was performed using the SSPACE software (Boetzer et al. 2011).
The RAST (Aziz et al. 2008) pipeline was used through its web interface to provide annotation for the assembled genomes, using the L. monocytogenes EGD-e genome (NC_003210) as reference.
PFAM domains were annotated to predict protein-coding genes with the PfamScan program (Mistry et al. 2007), using both the Pfam-A and Pfam-B domain models.
Gene Ontology (GO) terms were annotated to protein-coding genes directly from the Pfam domains annotation, using a custom script and the Pfam2go file available on the GO consortium website (http://ftp.cbi.pku.edu.cn/pub/database/GO/goa/external2go/pfam2go, last accessed March 10, 2015).
Clusters of Orthologous Genes
All-against-all BLASTP (Altschul et al. 1990) was performed using the BLOSUM80 matrix and accepting only best reciprocal hits with e value ≤1e-5 and where “second-best” hits from the same genome produce bit scores less than 90% of that associated with the best match. Putative clusters of orthologous genes (COGs) were established as groups of best reciprocal BLAST (Basic Local Alignment Search Tool) hits. Core genes were defined as COGs containing single representatives from all genomes considered (or all genomes within major clades), and accessory genes as COGs with incomplete representation.
Rarefaction Analyses of Core and Accessory Genomes
For each number of organisms considered (2–27 for Listeria sensu lato, 2–10 for Listeria sensu strictu), the inferred sizes of core and accessory genomes were recorded for 10,000 replicates of randomly selected combinations of genomes. Plots were prepared showing mean and standard deviation of these statistics.
Prunier
Prunier (version 2.1) was used with standard parameters, and in conjunction with RAXML version (7.2.6) for the estimation of the phylogenetic trees.
Phyletic Patterns and Inference of Gene Gains/Losses
The phyletic pattern of COGs inferred not to contain xenologous sequences was analyzed with the GLOOME program (version 1.266), using the default parameters, other than setting the optimization level to “high” and requesting reconstruction of ancestral state and generation of a gene absence/presence tree.
Genes were considered to be ancestral to a given node if the posterior probability of their presence, according to GLOOME, was ≥95%. Genes gains were assigned to the branch with the highest gain probability, provided that the presence probability at the ancestral node was ≤75% and ≥95% at the derived node.
dN/dS ratios were estimated for concatenated alignments of 623 genes universally present in Brochothrix and Listeria genomes using the Ka/Ks calculator (Zhang et al. 2006) with default settings.
Validation COGs Supporting LGT and Characterization of Genes Gained According to GLOOME
COGs supporting an LGT event according to Prunier and COGs consisting of less than five genes were validated by executing BLAST searches against an extensive database containing all the protein-coding sequences from all the 1,743 complete bacterial genomes in GenBank (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/all.gbk.tar.gz, last accessed March 10, 2015) for a total of 4,902,485 bacterial proteins.
Only hits showing an e value ≤1e-5 and covering the query sequence by more than 25% of its length were considered.
COGs were considered valid if and only if all the members remained best reciprocal BLAST hits among themselves even when searched for similarity against this more extensive database.
Similarly the genes gained at each node according to GLOOME were subjected to BLAST searches against the same database and using the same cut-offs, to discern whether they were more likely genes acquired by LGT (best matches outside Listeria) or generated by gene duplication (best matches within the same major Listeria group)
Phylogenetic Analyses
Phylogenetic analyses were performed on the concatenated alignment of a reference set of 623 proteins, conserved among the genomes of Listeria, Brochothrix, and the selected outgroups.
Peptide sequences for each gene were aligned individually with the Muscle software (Edgar 2004); conserved alignment blocks were identified and extracted using Gblocks (Castresana 2000) and concatenated using a custom Perl script.
Different amino acid substitution models (WAG [Whelan and Goldman], LG, JTT [Jones, Taylor, and Thorton], and Dayhoff) were compared using the ProtTest program (Darriba et al. 2011).
Phylogenetic relationships and bootstrap proportions were estimated using PhyML (Guindon et al. 2009) under the WAG amino acid substitution model (Whelan and Goldman 2001) with invariable and four gamma-distributed substitution rate categories.
Splits tree was constructed using the filtered supernetwork algorithm as implemented in Splitstree (Huson and Bryant 2006) with MinNumberTrees = 10 and default parameters, starting from the individual genes trees of the 623 Listeria and Brochothrix core genes.
Identification of Orthologous Genes of the Listeria Eut–Cob–Pdu and Flagellar Clusters
The sequences of protein-coding genes of the eut–cob–pdu and flagellar clusters were manually extracted from the NCBI (National Center for Biotechnology Information) entry of the Listeria monocytogenes EGD-e (AL591824) genome and searched against a custom protein database consisting of 4,902,485 bacterial proteins from 1,743 bacterial genomes, downloaded from ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/all.gbk.tar.gz. Only hits showing an e value ≤1e-5 and covering the query sequence by more than 25% of their length were considered. If at least 80% of the genes in a given cluster recovered satisfactory matches against proteins from a particular genome, we considered the cluster to be present in that genome and used the putative orthologs in the reconstruction of the phylogeny of the cluster. To avoid redundancies, we included only a maximum of two representative genomes for each species.
Consensus Networks
Consensus networks were calculated using the galled consensus network algorithm implemented in Dendroscope (Huson et al. 2009), starting from the individual genes trees and using a threshold of 20%.
Functional Enrichment Analyses
Functional enrichment analyses were performed using a custom script, implementing a hypergeometric test with a Bonferroni correction, the pan genome was used to provide a background for functional enrichment testing
Analysis of Pfam Domains Enrichment by Major Listeria Clade
The number of occurrences of each Pfam domain in each major clade (Listeria sensu strictu, Listeria grayi, Listeria fleischmanii, and Listeria rocourtiae) was counted using a custom script. A simple R script based on the hypergeometric distribution and implementing a Bonferroni correction was used to compute the P value for the overrepresentation of the domains in each group.
All the significantly enriched domains were included in the heatmap, which we computed using the heatmap.2 function of the gplots package in R.
Results
Preliminary Characterization of Two Novel Listeria-Like Isolates from Southern Italy
Two Listeria-like bacterial strains were isolated during a microbiological study of milk and milk-derived products in southern Italy. The sequence of a 1,473-bp fragment of 16S rDNA from a Listeria-like bacterium isolated from cheese samples was identical to a 16S rRNA sequence from L. fleischmannii in GenBank (e.g., accession number: JN093102) and the isolate was provisionally classified as a L. fleischmannii strain, a conclusion supported by metabolic and antibiotic resistance profiling (supplementary methods and results and tables S1 and S2, Supplementary Material online) as well as by subsequent genome sequencing and analysis (see below).
For a second isolate, from raw milk, a partial (1,388 bp) 16S rDNA sequence showed greater than 99% identity to L. rocourtiae and L. newyorkensis homologs in GenBank. However, metabolic profiles of our isolate and that of L. newyorkensis (Weller et al. 2014) (supplementary methods and results and tables S1 and S2, Supplementary Material online) did not conclusively resolve this issue. Assignment of our isolate to L. newyorkensis was achieved through genome sequencing and phylogenetic analyses (see below).
Genome Sequencing, Assembly, and Annotation
Listeria fleischmannii
In total, 1,341,131 pair-end reads of 250-bp were produced from L. fleischmannii genomic DNA using the Illumina MiSeq platform. The average insert size was estimated to be around 723 bp. After applying strict quality filtering and read trimming, 948,303 high quality mate pairs reads, with average read length of 82.12 bp were retained.
The genome was assembled using Velvet (Zerbino and Birney 2008). Optimal K-mer size and K-mer coverage parameters were established using a simple grid search (values of K from 21 to 81 bp) coupled with careful examination of K-mer frequency plots.
The final assembly (K = 53, expected coverage = 38×), contained 40 contigs longer than 500 bp with an N50 value of 119,961 bp. The estimated genome size was 2,842,057 bp. Scaffolding reduced the 40 contigs into 18 scaffolds and increased the N50 to 237,103 bp.
The RAST pipeline (Aziz et al. 2008) predicted 2,881 protein-coding genes, consistent with the 2,835–2,923 reported for other L. fleischmannii strains (Bertsch et al. 2012; den Bakker et al. 2013, 2014). A total of 5,008 Pfam domains, associated with 6,408 GO terms, were annotated to the protein-coding genes using the PfamScan program (Mistry et al. 2007) with default parameters. The draft genome sequence of the novel L. fleischmannii isolate is deposited in GenBank under the accession number AZHO00000000, the version described in this article is AZHO01000000.
Listeria newyorkensis
In total, 1,596,258 pair-end reads of 250 bp were produced from genomic DNA isolated from the L. newyorkensis isolate using the Illumina MiSeq platform with average insert size estimated at 650 bp. In total, 1,003,123 high quality pairs (mean read length 75.6 bp) were used as described above for genome assembly.
The final assembly (K = 61, expected coverage = 38×) contained 113 contigs of 500 bp or more. The estimated genome size was 3,321,213 bp and the N50 89,213 bp. Scaffolding merged the 113 contigs into 79 scaffolds and increased the N50 to 105,891 bp.
RAST identified 3,268 genes in the genome assembly, PfamScan found 5,556 Pfam domains in the predicted protein-coding genes, corresponding to 6,981 GO terms. The assembled contigs showed an Average Nucleotide Identity (Konstantinidis and Tiedje 2005) score of 99% with L. newyorkensis genomes, supporting the assignment of our isolate to this species. The draft genome sequence of the novel L. newyorkensis isolate is deposited in GenBank under the accession number AZHN00000000, the version described in this article is AZHN01000000.
Repetitive Sequences
Due to the relatively fragmented nature of the draft genome assemblies, it is difficult to distinguish confidently between plasmid borne sequences and repetitive elements in the main chromosomes. Consideration of the K-mer coverage of contigs in both assemblies revealed a restricted number of relatively short contigs with atypical coverage. For L. newyorkensis, contigs representing small and large subunit ribosomal DNA sequences had K-mer coverage of 328 and 385, respectively, with the median K-mer coverage of all contigs being 55, consistent with the presence of 6 or 7 copies of the ribosomal repeat. Three contigs (500–1,400 nt), estimated as showing copy numbers between 3 and 12, potentially encode proteins with high similarity to transposases/integrases, whereas another (copy number = 3–4) might encode a homolog of an LPXTG-motif protein cell wall anchor domain protein and a contig of approximately 900 nt (copy number = 2–3) potentially encodes an autolysin modifying protein. Taken together, we interpret these findings as indicating the presence of a number of possibly active transposable elements in the L. newyorkensis genome.
For L. fleischmannii, the median K-mer coverage of all contigs was 42.5, whereas ssu and lsu rDNA contigs had coverage of 263 and 290, respectively, again indicating the presence of 6 or 7 copies of the ribosomal repeat. No other contigs with anomalous coverage were detected and, taken together with the lower degree of fragmentation in this assembly, we suggest that other perfectly repeated, or high copy number sequences, such as transposons, are scarce or absent from the L. fleischmannii genome although it is difficult to exclude the presence of low copy number plasmid(s) in either organism.
Phylogeny of the Genus Listeria Sensu Lato
The genomes of 25 Listeria and 2 Brochothrix isolates (B. campestris and B. thermospacta) (supplementary table S3, Supplementary Material online), were selected on the base of phylogenetic relevance and contiguity of their genome assembly, downloaded from GenBank, and used in conjunction with our newly assembled genomes for comparative genomic analyses.
Clusters of reciprocal best BLASTP matches (putative COGs) were established using all-against-all BLASTP (Altschul et al. 1990) searches employing the BLOSUM80 matrix and accepting only best reciprocal hits with e value ≤ 1e-5 and where “second-best” hits produce bit scores less than 90% of that associated with the best match.
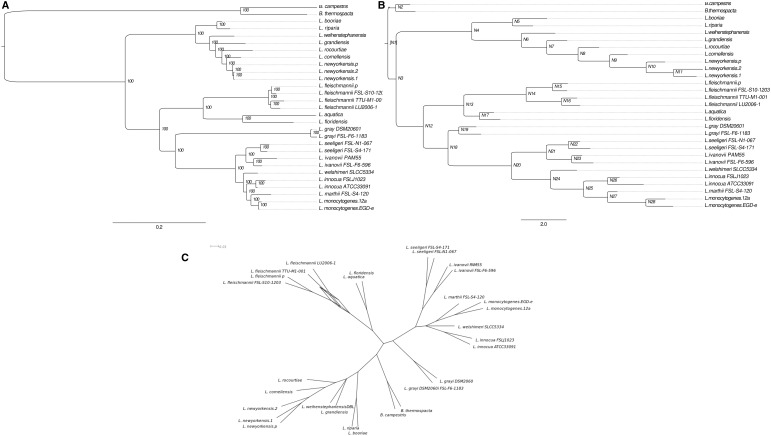
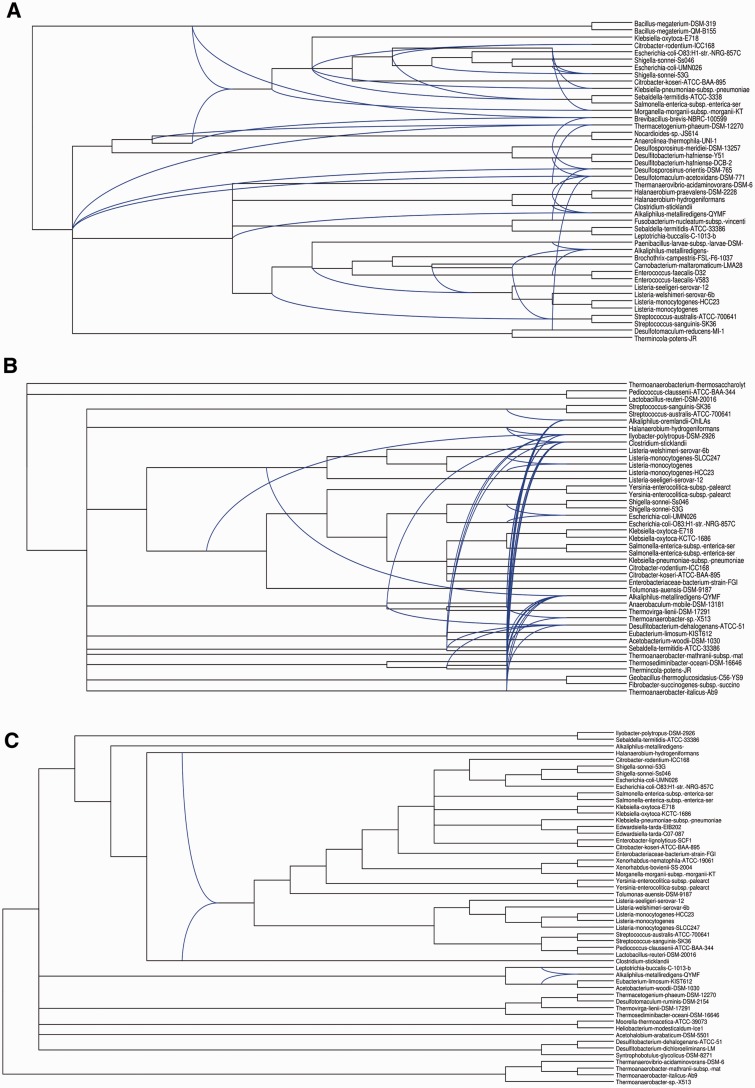
The conceptually translated sequences of 623 Listeria core genes also present in both Brochothrix genomes were independently aligned using Muscle (Edgar 2004) and ambiguously aligned regions were excluded using the GBlocks software (Castresana 2000). Maximum-likelihood phylogenetic reconstruction and bootstrap analyses were performed using the software PHYML (Guindon et al. 2009) under the WAG (Whelan and Goldman 2001) substitution model (suggested by the software ProtTest [Darriba et al. 2011] to best fit the data) with invariable and four gamma-distributed substitution rate categories. The resulting topology (fig. 1A) is highly congruent with that previously published by Weller et al. (2014) and further confirms the preliminary species assignments of the novel isolates sequenced here. All L. fleischmannii isolates were monophyletic and form a sister group to L. floridensis and L. aquatica as previously proposed (den Bakker et al. 2014). Listeria newyorkensis emerged as part of a well-supported monophyletic group containing L. rocourtiae, L. cornellensis, L. riparia, L. grandensis and L. weihenstephanensis, but not as sister to L. rocourtiae itself. This last grouping constituted the basal emergence within Listeria (100% bootstrap support). As expected, L. grayi represents the sister taxon of Listeria sensu strictu. The filtered supernetwork algorithm (Huson et al. 2004), as implemented in SplitsTree (Huson and Bryant 2006), was employed to provide a representation of possible conflicts between individual core gene trees (fig. 1C ) and provides no evidence for extensive lateral gene transfer/gene replacement/allele sorting of Listeria core genes other than, possibly between L. fleischmannii isolates. Manual observation of 60 individual gene trees conflicting with the concatenated gene tree revealed consistently low bootstrap support for noncanonical relationships between L. fleischmannii isolates. Taken together, these observations confirm existing hypotheses regarding phylogenetic relationships among Listeria sensu lato and suggest that lateral transfer/homologous replacement of genes that are widely distributed among Listeria sensu lato is rare.
Fig. 1.
—Phylogenetic relationships within the genus Listeria sensu lato. (A) Phylogenetic relationships inferred from maximum-likelihood analysis of a concatenation of 623 genes constituting the empirically observed core genome of Brochothrix and Listeria. Bootstrap proportions (100 replicates) are shown. (B) Gene presence/absence tree (8,211 genes present in at least 2 Listeria genomes) inferred using GLOOME. Node numbers are those referred to in the main text and tables whereas branch-lengths represent ratios of inferred gene gain to gene loss rates. (C) Splitstree consensus representation of 623 individual gene trees from the empirically observed core genome of Brochothrix and Listeria.
Core and Accessory Genomes
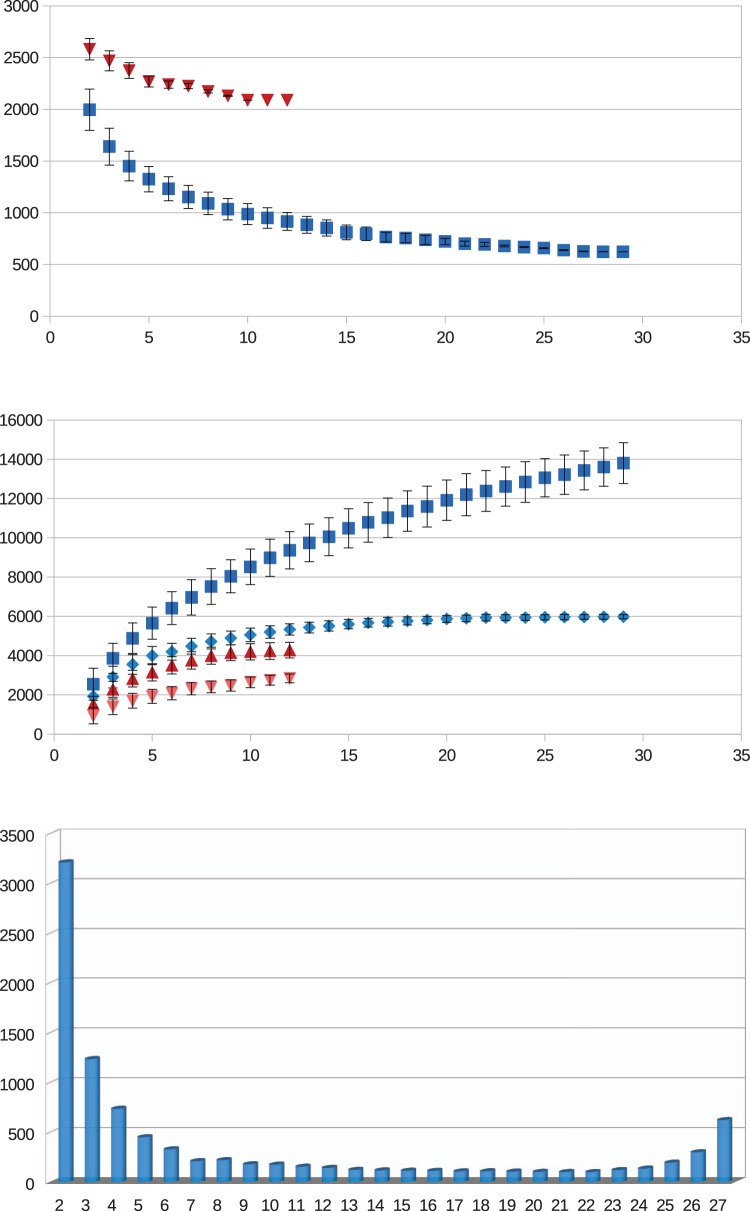
Our approach based on best reciprocal BLAST hits identified a total of 9,639 clusters (more than one gene) of putative orthologs as well as 9,095 singleton genes. Of the former 1,129 are present in all Listeria genomes and 2,086 in all Listeria sensu strictu genomes sampled. The latter observation was consistent with a previous empirical observation of a core genome of Listeria sensu strictu of 2,032 genes and a predicted size of 1,994 genes (den Bakker, Cummings et al. 2010). Rarefaction analyses suggested that these estimates of the core genomes of Listeria sensu strictu and Listeria sensu lato should remain fairly stable with additional species sampling (fig. 2A).
Fig. 2.
—Estimation of completeness of core and accessory genomes by rarefaction analyses. Core and accessory genome sizes were calculated on randomly resampled combinations of Listeria genomes. Trends according to numbers of genomes sampled are presented. Error bars represent standard deviations over 10,000 experimental replicates. (A) Core genome rarefactions for Listeria sensu strictu (red triangles) and the complete genus Listeria sensu lato (blue squares). (B) Accessory genome rarefactions for Listeria sensu strictu without singleton genes (light red triangles), with singleton genes (dark red triangles) and Listeria sensu lato without singleton genes (light blue diamonds) with singleton genes (dark blue squares). (C) Frequency distribution of number of entries in inferred COGS for all Listeria genomes used in this study.
Analogously, absolute counts of the accessory genome of Listeria sensu strictu and sensu lato were 4,280 and 13,805 genes, respectively. Rarefaction analyses suggest that the catalog of the accessory genes of Listeria sensu strictu is fairly completely represented by the sampling used and irrespective of the inclusion of singleton genes, whereas that of Listeria sensu lato appears close to complete when singleton genes are excluded, but rather open when singleton genes are included (fig. 2B). The distribution of frequency of occurrence of genes among Listeria genomes (fig. 2C) confirms that a large proportion of accessory genes are present in only one or two genomes. The discrepancy between the observed pan genome size of Listeria sensu strictu (4,280 genes) and a previous total (2,918 genes) (den Bakker, Cummings et al. 2010) is due mostly to the inclusion of genes with less than 75 codons in this study. Boxplots of gene size distributions by genome are presented in supplementary figure S1, Supplementary Material online.
Lateral Gene Transfer within Listeria
Prunier (Abby et al. 2010) searches for potential lateral gene transfer events by identifying gene/protein trees that show well-supported incongruence with a reference organismal tree that has been pruned to give identical organismal sampling as the gene under examination. This software was used to examine 4,089 clusters of best reciprocal BLAST hits containing representatives from five or more genomes. The possible confounding effect of LGT on COG assignment has recently been noted (Dalquen et al. 2013). Indeed, of 738 candidate transfers identified by Prunier, additional similarity searches (see Materials and Methods) suggested that 416 were likely “xeno-COGs” wherein one or more member(s) were derived from homologous gene replacement or independent acquisition events involving donors from outside the genus Listeria. COGs whose phylogenies were identified by Prunier as incongruent are listed in supplementary table S4, Supplementary Material online. Notably, none of the 60 core loci whose poorly supported phylogenies had suggested possible transfers between L. fleischmannii isolates was recovered by Prunier as showing statistically significant incongruence.
The high frequency of inferred xeno-COGs among clusters with five or more genes prompted an evaluation—through similarity searches—of the coherence of 5,500 COGs that contained representatives from only 2 to 4 genomes (and which were accordingly not suitable for analysis by Prunier). Of these, 1,012—where at least one member of the cluster recovered a best BLAST match outside Listeria—were inferred to be xeno-COGs. In total, 386 clusters with incomplete representation in more than one Listeria subclade but where all members were more similar to each other than to nonlisterial genes were considered candidates for within Listeria gene transfer.
Genome-Wide Evolutionary Dynamics
Presence/absence profiles of 8,211 putative orthologs, present in at least two genomes and for which no evidence of xenologous components had been inferred, were provided as input to GLOOME (Cohen et al. 2010) for reconstruction of a presence/absence phylogenetic tree, and to provide estimates of patterns of gene gain and loss across the genus. The gene presence–absence tree (fig. 1B) is congruent with the topology derived from likelihood analyses of concatenated gene sets (fig. 1A). GLOOME provides probabilities for gain or loss of each gene on each branch as well as posterior probabilities for the presence of each gene at each node in the tree. Gene gains were assigned to the most probable branch—provided that the presence probability at the following node was ≥95%. Singleton clusters were assigned as gains on terminal branches and the previously identified xeno-COGs were excluded from subsequent analyses.
The potential origins of genes identified by GLOOME as acquired along internal branches of the Listeria phylogeny and of “singleton” best BLAST match clusters (genes present in a single Listeria genome) were further investigated through similarity searches (see Materials and Methods). Amino acid sequences yielding best matches to proteins already included in Listeria putative COGS were tentatively assigned as resulting from Listeria-specific gene duplication events, whereas those generating best matches to non-Listeria proteins were considered candidates for acquisition by lateral gene transfer.
Of 2,593 genes identified as acquisitions at the base of, or within, the L. fleischmannii clade, 997 provided significant best BLAST matches outside Listeria. For the L. rocourtiae clade, the equivalent figures were 3,941 and 1,524; for L. grayii 549 and 285 and for Listeria sensu strictu of 2,432 inferred nonterminal branch acquisitions 1,445 recovered best hits outside Listeria. Although ratios of apparent gains by LGT and duplication remain fairly constant across the genus, the proportion of gains assigned to duplication events is rather higher on terminal branches (54%) than internal branches (33%). Statistics regarding the numbers and nature of inferred gene gain events in the distinct Listeria subclades are summarized in table 1.
Table 1.
Statistics regarding Gene Gain and Loss Rates for Branches within the Listerial Phylogeny as Inferred by GLOOME and Singleton Assignment to Terminal Branches
| Branch | Lengtha | Gainsb | Lossesb | Gainc | Lossc | Ratiod |
|---|---|---|---|---|---|---|
| N2 | 0.46 | 160 | 461 | 2.86E-003 | 9.92E-004 | 0.35 |
| Brochothrix campestris | 0.16 | 196 | 308 | 8.40E-004 | 5.36E-004 | 0.64 |
| Brochothrix thermospacta | 0.14 | 232 | 268 | 6.17E-004 | 5.35E-004 | 0.87 |
| N3 | 0.31 | 469 | 170 | 6.65E-004 | 1.84E-003 | 0.36 |
| N4 | 0.15 | 538 | 161 | 2.81E-004 | 9.39E-004 | 3.35 |
| N5 | 0.05 | 251 | 140 | 1.88E-004 | 3.36E-004 | 1.79 |
| Listeria booriae | 0.01 | 279 | 183 | 3.86E-005 | 5.88E-005 | 1.52 |
| Listeria riparia | 0.03 | 284 | 313 | 1.00E-004 | 9.09E-005 | 0.91 |
| N6 | 0.03 | 286 | 131 | 1.08E-004 | 2.37E-004 | 2.19 |
| Listeria weihenstephanensis | 0.05 | 350 | 234 | 1.53E-004 | 2.29E-004 | 1.50 |
| N7 | 0.01 | 232 | 207 | 5.60E-005 | 6.30E-005 | 1.12 |
| Listeria grandiensis | 0.06 | 285 | 318 | 2.27E-004 | 2.04E-004 | 0.90 |
| N8 | 0.01 | 238 | 172 | 5.89E-005 | 8.14E-005 | 1.38 |
| Listeria rocourtiae | 0.07 | 305 | 330 | 2.17E-004 | 2.00E-004 | 0.92 |
| N9 | 0.01 | 251 | 181 | 3.98E-005 | 5.54E-005 | 1.39 |
| Listeria cornellensis | 0.03 | 281 | 354 | 1.10E-004 | 8.78E-005 | 0.79 |
| N10 | 0.01 | 346 | 214 | 4.05E-005 | 6.54E-005 | 1.62 |
| Listeria newyorkensis.p | 0.00 | 134 | 160 | 8.23E-006 | 6.87E-006 | 0.83 |
| N11 | 0.00 | 173 | 143 | 1.12E-005 | 1.35E-005 | 1.21 |
| Listeria newyorkensis.2 | 0.00 | 150 | 116 | 1.03E-005 | 1.33E-005 | 1.29 |
| Listeria newyorkensis.1 | 0.00 | 125 | 123 | 1.31E-005 | 1.33E-005 | 1.02 |
| N12 | 0.07 | 244 | 203 | 3.04E-004 | 3.64E-004 | 1.20 |
| N13 | 0.09 | 311 | 173 | 3.02E-004 | 5.44E-004 | 1.80 |
| N14 | 0.14 | 437 | 159 | 3.23E-004 | 8.87E-004 | 2.75 |
| N15 | 0.01 | 155 | 133 | 4.31E-005 | 5.03E-005 | 1.17 |
| Listeria fleischmannii.p | 0.00 | 78 | 116 | 4.65E-006 | 3.12E-006 | 0.67 |
| Listeria fleischmannii FSL-S10 | 0.01 | 124 | 195 | 8.01E-005 | 5.12E-005 | 0.64 |
| N16 | 0.01 | 162 | 117 | 3.11E-005 | 4.31E-005 | 1.39 |
| Listeria fleischmannii TTU-M1 | 0.03 | 216 | 315 | 1.32E-004 | 9.08E-005 | 0.69 |
| Listeria fleischmannii LU2006 | 0.02 | 387 | 466 | 5.22E-005 | 4.33E-005 | 0.83 |
| N17 | 0.09 | 220 | 320 | 3.87E-004 | 2.66E-004 | 0.69 |
| Listeria aquatica | 0.16 | 302 | 480 | 5.20E-004 | 3.28E-004 | 0.63 |
| Listeria floridensis | 0.11 | 342 | 373 | 3.20E-004 | 2.94E-004 | 0.92 |
| N18 | 0.03 | 230 | 220 | 1.48E-004 | 1.55E-004 | 1.05 |
| N19 | 0.29 | 235 | 492 | 1.24E-003 | 5.93E-004 | 0.48 |
| Listeria gray DSM20601 | 0.00 | 145 | 189 | 1.85E-005 | 1.42E-005 | 0.77 |
| Listeria grayi FSL-F6-1183 | 0.01 | 221 | 226 | 6.08E-005 | 5.96E-005 | 0.98 |
| N20 | 0.13 | 510 | 182 | 2.55E-004 | 7.13E-004 | 2.80 |
| N21 | 0.02 | 189 | 122 | 1.22E-004 | 1.88E-004 | 1.54 |
| N22 | 0.03 | 194 | 199 | 1.65E-004 | 1.61E-004 | 0.97 |
| Listeria seeligeri FSL-N1-067 | 0.02 | 194 | 381 | 9.58E-005 | 4.86E-005 | 0.51 |
| Listeria seeligeri FSL-S4-171 | 0.01 | 247 | 256 | 5.75E-005 | 5.56E-005 | 0.97 |
| N23 | 0.02 | 148 | 132 | 1.08E-004 | 1.21E-004 | 1.12 |
| Listeria ivanovii PAM55 | 0.05 | 266 | 578 | 1.88E-004 | 8.67E-005 | 0.46 |
| Listeria ivanovii FSL-F6-596 | 0.02 | 238 | 249 | 7.74E-005 | 7.40E-005 | 0.96 |
| N24 | 0.02 | 186 | 107 | 9.15E-005 | 1.59E-004 | 1.74 |
| Listeria welshimeri | 0.03 | 195 | 169 | 1.59E-004 | 1.83E-004 | 1.15 |
| N25 | 0.01 | 169 | 146 | 4.11E-005 | 4.76E-005 | 1.16 |
| N26 | 0.02 | 107 | 100 | 1.88E-004 | 2.00E-004 | 1.06 |
| Listeria innocua ATCC33091 | 0.02 | 202 | 390 | 1.02E-004 | 5.27E-005 | 0.52 |
| Listeria innocua FSLJ1023 | 0.00 | 204 | 129 | 1.68E-005 | 2.66E-005 | 1.58 |
| N27 | 0.01 | 121 | 119 | 8.25E-005 | 8.39E-005 | 1.02 |
| Listeria marthii FSL-S4-120 | 0.04 | 203 | 477 | 2.13E-004 | 9.06E-005 | 0.43 |
| N28 | 0.01 | 235 | 139 | 4.05E-005 | 6.83E-005 | 1.69 |
| Listeria monocytogenes.12a | 0.01 | 120 | 141 | 5.61E-005 | 4.80E-005 | 0.86 |
| Listeria monocytogenes.EGD-e | 0.00 | 113 | 95 | 1.79E-005 | 2.15E-005 | 1.20 |
aBranch length (substitutions per site from concatenated core gene phylogeny).
bExpected number of gene gains/losses according to GLOOME.
cSubstitutions per gene gain/loss.
dRatio of gene gain/loss rates.
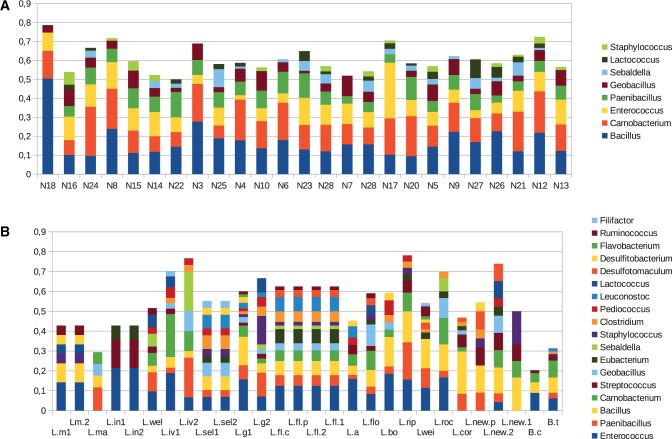
Perhaps unsurprisingly, the largest numbers of genes were gained on the branches leading to the basal divergences within each of the major subclades within Listeria. Considering all acquisitions assigned to branches associated with each major Listeria clade, no obvious biases toward particular possible donor organisms were evident (fig. 3) although for all clades the most frequently inferred potential donors included Bacillus, Carnobacterium, Entrococcus, and Paenibacillus.
Fig. 3.
—Best matches for genes inferred to have been gained by horizontal transfer from outside Listeria. Frequencies of best matches to distinct taxa for genes inferred to have been acquired on specific branches and giving best matches outside Listeria. All entries correspond to branches leading to nodes described in table 1. (A) Genes inferred by GLOOME to have been inferred on internal branches of the Listeria phylogeny. (B) Singleton genes inferred to have been gained on terminal branches. Key: L.m1, L. monocytogenes 1/2a; L.m2, L. monocytogenes.EGD-e; L.ma, L. marthii FSL-S4-120; L.in1, L. innocua ATCC33091; L.in2, L. innocua FSLJ1023; L.wel, L. welshimeri SLCC5334; L.iv1, L. ivanovii PAM55; L.iv2, L. ivanovii FSL-F6-596; L.sel1, L. seeligeri FSL-S4-171; L.sel2, L. seeligeri FSL-S4-171; L.g1, L. grayi FSL-F6-1183; L.g2, L. grayi FSL-F6-1183; L.fl.c, L. fleischmannii TTU-M1; L.fl.p, L. fleischmannii this study; L.fl.2, L. fleischmannii LU2006-1; L.fl.1, L. fleischmannii FSL-S10-1203; L.a, L. aquatica; L.flo, L. floridensis; L.bo, L. booriae; L.rip, L. riparia; Lwei, L. weihenstephanensis; L.roc, L. rocourtiae; L.cor, L. cornellensis; L.new.p, L. newyorkensis this study; L.new.2, L. newyorkensis 2; L.new.1, L. newyorkensis 1; B.c, B. campestris; B.t, B. thermospacta.
Strikingly, the mean ratio of branch-specific rate of gene gain to rate of gene loss (defined as the sum of gene gain or loss probabilities assigned to a branch by GLOOME [table 2]) was significantly higher (1.4 vs. 1.1) in the basal (L. rocourtiae) clade than in the remaining clades (P ≤ 0.0171 Welch’s t-test). Normalization of the inferred branch-specific gene gain and loss rates to the lengths of branches in the concatenated gene tree (to provide estimates of rates of gene gain and loss with respect to fixed substitutions) (table 2) suggested that although rates of gene loss were similar across the genus, gene gains occur more rapidly in the basal clade (P = 6.097E-07), consistent with observations regarding genome size and gene complement across the Listeria tree (supplementary table S3, Supplementary Material online).
Table 2.
Summary of Specific Gene Gains for Individual Branches within the Listerial Phylogeny as Inferred from GLOOME Outputs and Singleton Assignment to Terminal Branches
| Branch | Gainsa | With Matchesb | Matches within SMCc |
|---|---|---|---|
| Brochothrix campestris | 495 | 446 | 236 |
| Brochothrix thermospacta | 505 | 489 | 218 |
| N3 | 403 | 357 | 61 |
| N4 | 390 | 364 | 87 |
| N5 | 301 | 289 | 129 |
| Listeria booriae | 461 | 222 | 195 |
| Listeria riparia | 396 | 324 | 290 |
| N6 | 151 | 127 | 59 |
| Listeria weihenstephanensis | 282 | 269 | 208 |
| N7 | 178 | 164 | 62 |
| Listeria grandiensis | 292 | 292 | 261 |
| N8 | 141 | 131 | 48 |
| Listeria rocourtiae | 397 | 342 | 310 |
| N9 | 196 | 168 | 79 |
| Listeria cornellensis | 496 | 447 | 389 |
| N10 | 268 | 393 | 189 |
| Listeria newyorkensis.p | 91 | 82 | 60 |
| N11 | 118 | 216 | 111 |
| Listeria newyorkensis.1 | 129 | 81 | 56 |
| Listeria newyorkensis.2 | 112 | 84 | 61 |
| N12 | 111 | 100 | 31 |
| N13 | 170 | 163 | 61 |
| N14 | 229 | 205 | 96 |
| N15 | 312 | 269 | 98 |
| Listeria fleischmannii.p | 149 | 95 | 85 |
| Listeria fleischmannii FSL-S10-1203 | 207 | 124 | 122 |
| N16 | 171 | 141 | 51 |
| Listeria fleischmannii TTU-M1 | 213 | 148 | 132 |
| Listeria fleischmannii LU2006-1 | 623 | 511 | 368 |
| N17 | 179 | 137 | 65 |
| Listeria aquatica | 454 | 385 | 279 |
| Listeria floridensis | 405 | 399 | 336 |
| N18 | 124 | 104 | 21 |
| N19 | 254 | 202 | 79 |
| Listeria gray DSM20601 | 235 | 198 | 156 |
| Listeria grayi FSL-F6-1183 | 180 | 111 | 69 |
| N20 | 386 | 379 | 77 |
| N21 | 60 | 58 | 19 |
| N22 | 208 | 181 | 69 |
| Listeria seeligeri FSL-N1-067 | 402 | 338 | 278 |
| Listeria seeligeri FSL-S4-171 | 375 | 314 | 254 |
| N23 | 181 | 163 | 59 |
| Listeria ivanovii PAM55 | 422 | 350 | 283 |
| Listeria ivanovii FSL-F6-596 | 375 | 357 | 187 |
| N24 | 209 | 202 | 67 |
| Listeria welshimeri SLCC5334 | 99 | 77 | 48 |
| N25 | 101 | 98 | 33 |
| N26 | 64 | 59 | 21 |
| Listeria innocua ATCC33091 | 238 | 200 | 153 |
| Listeria innocua FSLJ1023 | 125 | 41 | 27 |
| N27 | 42 | 38 | 13 |
| Listeria marthii FSL-S4-120 | 128 | 124 | 107 |
| N28 | 234 | 201 | 59 |
| Listeria monocytogenes.12a | 84 | 83 | 66 |
| Listeria monocytogenes.EGD-e | 113.2 | 93 | 72 |
aNumber of genes gained.
bNumber of gained genes with matches to nonredundant database.
cNumber of matches within the same major clade of Listeria.
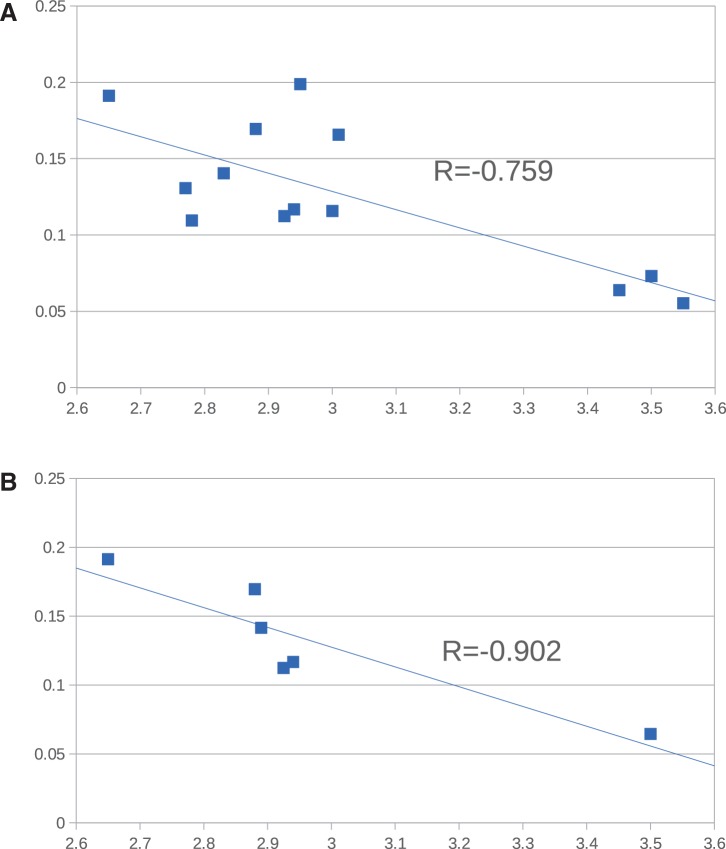
In the presence of purifying selection on coding sequences, higher dN/dS indicates a lower efficiency of selection and therefore a larger impact of drift, accordingly, dN/dS ratios derived from concatenated nucleotide alignments of the core genes used in phylogenetic reconstructions were employed as proxies of rates of genetic drift (Novichkov et al. 2009; Joseph et al. 2012) between pairs of cospecific isolates from each major clade within the genus Listeria. For the four L. fleischmanni and three L. newyorkensis genomes, all pairwise comparisons were performed whereas the two representatives of L. seeligeri, L. monocytogenes, L. ivanovii and L. innocua employed for GLOOME analysis were also compared. Estimated dN/dS values showed a significant negative correlation with genome size (r = −0.759, P = 0.002595) (fig. 4A). When only a single intraspecific comparison was used for L. fleischmanni and L. newyorkensis, the correlation coefficient (r = −0.9) remained significant (P = 0.01381) (fig. 4B), consistent with the hypothesis that lower rates of genetic drift (Novichkov et al. 2009) might underlie the larger genome size of basal clade Listeria.
Fig. 4.
—Correlation between dN/dS for core genes and genome size for intraspecific comparisons within Listeria. Ratios of nonsynonymous to synonymous substitution rates were calculated for cospecific genomes from 623 Brochothrix/Listeria core genes and correlated with genome size. (A) Scatterplot including permutations of L. fleischmannii and L. newyorkensis intraspecies comparisons. (B) Scatterplot including single L. fleischmannii and L. newyorkensis intraspecies comparisons.
Functional Enrichment Analyses
COGs predicted by Prunier to have been subject to LGT within Listeria (both inter- and intraclade) showed most significant enrichment of the GO terms GO:0004478 (methionine adenosyltransferase-activity), GO:0006556, (S-adenosylmethionine-biosynthetic-process) (supplementary table S5, Supplementary Material online). Xeno-COGs recovered by Prunier were enriched in various carbohydrate metabolic processes, whereas sparsely distributed xeno-COGs with less than five members showed strong enrichment of GO:0015074 (DNA-integration), consistent with the inference of their transfer (supplementary table S6, Supplementary Material online).
PFAM domains showing statistically significant departure from random distribution between major listerial clades were identified using a custom script (supplementary fig. S2 and table S7, Supplementary Material online). This analysis highlights, for example, the expansion of internalin domain genes in Listeria sensu strictu, as well as the overrepresentation domains associated with cobalamin metabolism (cbi/cob), ethanolamine metabolism (eut), and flagellar motility.
Functional enrichment analyses performed on gene sets identified as gains on individual branches of the listerial phylogeny (supplementary table S8, Supplementary Material online) also highlighted several patterns that were highly consistent with the global PFAM domain enrichments. For example, terms related to ethanolamine and cobalamin metabolism were significantly enriched on the branch leading to Listeria sensu strictu, whereas terms relating to flagellar mobility were enriched on the branch leading to the common ancestor of L. grayi and Listeria sensu strictu. Three examples regarding the putative gain of multigene clusters are considered in the following sections.
Riboflavin Biosynthesis in L. grayi
The significant enrichment of gene gains with associated GO terms related to “riboflavin-biosynthetic-process” on the branch leading to the last common ancestor of L. grayi isolates was associated with four, genomically clustered, genes annotated as RibD, RibE, RibAB, and RibH and showing highest similarity with homologs from various firmicutes. The gene arrangement and the presence of an annotated FMN-sensitive riboswitch are conserved with a wide range of bacterial operons responsible for the biosynthesis of riboflavin, an essential precursor of FAD and FMN (Perkins and Pero 2001). These observations lead to the prediction that L. grayi, unlike L. monocytogenes (Welshimer 1963) (and presumably other members of the genus), should be capable of riboflavin biosynthesis.
Acquisition of Flagellar Genes
Forty-two of the 124 genes gained inferred to have been gained in a common ancestor of L. grayi and Listeria sensu strictu were associated with GO terms relating to flagellar biosynthesis. Strikingly these genes constitute a contiguous cluster on the genome of L. grayi and “sensu strictu” species. Phylogenetic analyses of individual protein sequences (supplementary fig. S3, Supplementary Material online, and see Neville et al. 2012) represented as a galled network in figure 5 reveal the consistent affinity of listerial flagellar-system proteins for their homologs from the B. cereus complex, where the corresponding genes are similarly clustered in a tight genomic interval. Notably B. cereus proteins do not show strong phylogenetic affinity with other firmicute flagellar genes including those of other Bacillus species. The shared peculiarities of listerial and B. cereus complex flagellar apparatus include the presence of common FliN/Y paralogs absent from other genomes (Neville et al. 2012).
Fig. 5.
—Galled consensus network for flagellar genes inferred to have been gained in a common ancestor of L. grayi and Listeria sensu strictu. Individual gene trees were estimated for components of representative homologous flagellar gene clusters from listerial and nonlisterial genomes using the maximum-likelihood method and a galled consensus network was calculated using the Dendroscope software.
These data are consistent with lateral gene transfer of the entire flagellar biosynthetic pathway between a common ancestor of L. grayi/Listeria sensu strictu and an ancestor of the B. cereus complex. Alternatively, an ancestral flagellar apparatus in B. cereus may have been replaced from a similar, as yet unknown, source to the donor of the listerial flagella. Flagellar genes have been associated with pathogenicity in both B. cereus-like organisms (Bouillaut et al. 2005; Ramarao and Lereclus 2006) and Listeria (Dons et al. 2004) underlining the possible relevance of the acquisition of flagella, before the divergence of L. grayi, in the evolution of pathogenicity in Listeria.
Genetics of Ethanolamine and Propane-2-Diol Utilization in Listeria Sensu Strictu
Another striking example of functional enrichment is observed among genes inferred to have been acquired in the common ancestor of Listeria sensu strictu, where GO terms related to cobalt transport, cobalamin metabolism, ethanolamine metabolism, and propanediol metabolism were highly enriched. Indeed, 82 of the 386 genes acquired are clustered in a single, 53-kb locus in Listeria sensu strictu, and constitute the annotated cobalamin (vitamin B12) biosynthetic cluster and the propane-2-diol and ethanolamine utilization clusters, whose regulation has recently been studied in detail and shown to be highly integrated (Mellin et al. 2013, 2014) and, at least in the case of the eut pathway, contribute significantly to virulence in L. monocytogenes (Mellin et al. 2014).
The genomes of basal Listeria contain a broadly conserved putative operon consisting of cobA, NirS, nirB, cysG, and cbiX genes as well as a distant locus homologous to the Bacillus hemA, hemC, hemD, hemB, hemL operon (Hansson et al. 1991). HemH, hemE, hemN, and hemY genes (Hansson and Hederstedt 1992) are scattered throughout their genomes, whereas cob–cbi operons are not present. These observations lead us to hypothesize that basal Listeria are capable, like Bacillus megaterium, of synthesizing uroporphyrinogen III, heme and siroheme from a glutamyl tRNA precursor, but are unable to produce cobalamin (B12) cofactors de novo (Raux et al. 2003; Layer et al. 2010). The conservation of the structure of the hemACDBL and the similarity of the cobA–cbiX locus to homologous loci in Bacillus, and other relatives of Listeria suggest that this state may be ancestral to the genus Listeria. Brochothrix genomes lack the cobA–cbiX locus (although they possess a highly divergent cobA gene which shows highest sequence similarity to homologs from Clostridium species and was likely acquired independently); however, all genes expected to be required for heme biosynthesis are present—potentially consistent with secondary loss of the ancestral siroheme biosynthetic pathway in Brochothrix.
The capacity to utilize ethanolamine and/or propanediol as sole carbon sources is, for many bacteria, a prerequisite for pathogenesis of vertebrates (Conner et al. 1998; Joseph et al. 2006; Klumpp and Fuchs 2007; Srikumar and Fuchs 2010). Cobalamin is required as a cofactor in both these metabolic pathways and in Listeria sensu strictu, 40 characterized lineage-specific genes are consistently clustered in a single 53-kb genomic locus, and organized in 3 (putative) operons: a pdu (propanediol utilization) operon, a eut (ethanolamine utilization) operon, and a near complete cob–cbi (cobalamin biosynthesis) operon (the last stage of synthesis of functional cobalamin cofactor is mediated by a noncanonical combination of enzymes [Gray and Escalante-Semerena 2010]). This gene cluster is flanked at one end by the ancestral cysG gene. Other genes from the putatively ancestral cobA, NirS, nirB, cysG, and cbiX cluster are not present in Listeria sensu strictu, presumably as they are not required for cobalamin or siroheme synthesis in the presence of a complete complement of cob–cbi genes. It was previously proposed that eut, pdu, and cob–cbi genes might have been acquired through horizontal gene transfer from a Salmonella-like bacterium (Buchrieser et al. 2003).
Consensus networks phylogenetic reconstruction for both individual (supplementary fig. S4, Supplementary Material online) pdu and cob–cbi genes and gene sets (fig. 6A and B) suggest a close phylogenetic affinity of Listeria pdu and cob–cbi genes to proteobacterial homologs as well as, in the case of cob–cbi to similarly arranged clusters in Gram-positive bacteria including Lactobacillus, Pediococcus, and Streptococcus. The galled networks suggest possible recombination or gene replacement within the listerial clusters, although uncertainties regarding the rooting of these representations render explicit inferences difficult.
Fig. 6.
—Galled consensus networks for pdu/cob-cbi and eut genes inferred to have been gained in a common ancestor of Listeria sensu strictu. Individual gene trees were estimated for components of representative homologous gene clusters from listerial and nonlisterial genomes using the maximum-likelihood method and galled consensus networks were calculated using the Dendroscope software. (A) Pdu cluster proteins, (B) Cob–cbi cluster proteins, (C) Eut cluster proteins.
Although Eut genes are also present in Brochothrix, neither phylogenetic reconstructions of individual eut genes (supplementary fig. S4, Supplementary Material online) nor the galled network representation (fig. 6C ) suggests a sister group relationship with the listerial genes, and unlike pdu and cob–cbi, no proteobacterial affinities were identified. Instead, the network consensus representation implies that the listerial eut cluster is made up of genes with firmicute ancestry.
Taken together, we interpret these data as suggesting that the eut, cob–cbi, and pdu operons have likely been acquired by Listeria through lateral gene transfer. Although the arrangement and content of the cob–cbi and pdu clusters are conserved with proteobacterial exemplars, at least in the case of cob–cbi, similar gene arrangements are also observed in a variety of firmicutes. Uncertainties regarding the root positions for gene trees make direct inferences regarding the nature of donors difficult. An alternative model, wherein this cluster was present in the common ancestor of Listeria and Brochothrix (with at least three independent losses in the known basal Listeria lineages), cannot be excluded. However, the resemblance of the Brochothrix eut cluster to that found in Enterococcus and Streptococcus and similarities between the Listeria and Clostridia eut clusters may indicate that Brochothrix and Listeria sensu strictu acquired the operon independently.
Discussion
Unraveling the molecular mechanisms underlying the pathogenicity of L. monocytogenes has been a major consideration motivating genome sequencing within the genus Listeria. Comparative genomics/transcriptomics studies (Schmid et al. 2005; Hain et al. 2006; den Bakker, Bundrant, et al. 2010; den Bakker, Cummings, et al. 2010; Wurtzel et al. 2012; Kuenne et al. 2013) largely agree that on several key points:
The common ancestor of Listeria sensu strictu was likely a pathogen, as many pathogenicity-associated genes are consistently found in nonpathogenic sensu strictu species and strains (Schmid et al. 2005).
The loss or attenuation of pathogenicity in several Listeria sensu strictu strains and species results from the loss or inactivation of key pathogenicity genes.
The core genome of Listeria sensu strictu consists of about 2,000 genes and is highly stable. Gene acquisition and loss occurs, but is rather limited (den Bakker, Cummings, et al. 2010).
In L. monocytogenes, the majority of gene scale differences occurs at a limited number of hypervariable genomic hot-spots (Kuenne et al. 2013).
In this work, we have characterized the genomes of two additional basal Listeria isolates and used comparative genomics approaches to investigate the genome-level evolutionary dynamics of the entire genus, with a focus on the “basal” Listeria which have been less intensively investigated. In the following paragraphs, the main inferences from our analyses will be considered and contextualized.
The phylogenetic relationships within Listeria sensu lato inferred here from a set of 623 proteins encoded by genes with orthologs in all Listeria and Brochothrix genomes are congruent with previous analyses (Weller et al. 2014) and show that the genus is made up of four subclades, wherein L. grayi is sister to Listeria sensu strictu. A clade consisting of L. fleischmannii, L. floridensis and L. aquatica is the sister of Listeria sensu strictu/L. grayi, whereas the remaining species constitute the basal clade in the genus. Notably, the mean genome size of members of the basal clade is significantly larger than that of the remaining congeners. For pairs of genomes from the same species, relative nonsynonymous to synonymous substitution rates in core genes were used as a proxy for levels of genetic drift (Novichkov et al. 2009; Joseph et al. 2012) and showed a significant correlation with genome size across pathogenic and nonpathogenic species. Similar observations have previously been made over a taxonomically wide sampling of bacteria and archaea (Novichkov et al. 2009). To our knowledge, this is the first such inference from multiple pairs of congeneric species.
For comparative genomic analyses regarding gene gain and loss in bacteria, the authors of a previous study in Listeria sensu strictu (den Bakker, Cummings et al. 2010) elected to exclude predicted open-reading frames (ORFs) encoding proteins shorter than 75 amino acids from analyses; due to potentially inconsistent annotation of such ORFs between genomes considered (Warren et al. 2010). There also exists a widespread suspicion that a high proportion of short ORFs might be prediction artifacts; however, a mounting body of evidence suggests that many, if not most, such ORFs are likely to represent real genes (Ochman 2002; Wang et al. 2008; Su et al. 2013), and that rapid turnover of short ORFs might represent a significant mechanism in prokaryotic genome evolution (Nowell et al. 2014). Here, we have chosen to retain all predictions for analyses. The principal consequence of overprediction of spurious genes is likely to be the inflation of numbers of singleton genes and COGS with few genomes represented, whereas inconsistent annotation will lead, in the main, to slight overestimation of gene loss frequencies.
Rarefaction analyses supported previous conclusions that the catalog of the core genome of Listeria sensu strictu is essentially complete (den Bakker, Cummings et al. 2010), and that a similar situation applies for Listeria sensu lato. However, the pan-genome of Listeria sensu lato is likely to expand rapidly with the addition of further genome sequences. As with other bacterial genera (e.g., Touchon et al. 2014), singleton genes constitute a notable component of the accessory genome and contribute greatly to estimates of its rate of expansion as a function of genome sampling. Nevertheless, even in the absence of singleton genes our analyses fail to suggest convergence of accessory genome size estimates, indicating that overprediction of genes is unlikely to completely explain this inference.
Probabilistic reconstruction of evolutionary relationships based on gene presence/absence recovered a topology which was consistent with molecular sequence data, whereas inferences of gene gain and loss rates across the genus suggested that both the ratio of gene gain to gene loss and absolute rates of gene gain (gain rate normalized to branch length in substitutions) were significantly higher in the basal (L. rocourtiae) clade than in remaining taxa. These observations are consistent with the observed differential genome sizes and dN/dS ratios and indicate that quite distinct dynamics apply to genome evolution in the basal clade.
Similar proportions of gene gains are inferred to result from duplication and acquisition by gene transfer across the genus, although duplication is more frequent among recent gains (singleton genes), consistent with the hypothesis that although transient duplication events may be relatively frequent, fixation of exogenous genetic is more probable than fixation of duplicated material. Perhaps unsurprisingly, putative sources of genes gained through lateral transfer were most frequently relatively closely related organisms from within the firmicutes. No bias toward particular candidate donors was observed for individual clades or genomes although sets of genes gained on several branches showed highly significant functional enrichments (see below).
A substantial number of LGT events between Listeria genomes were initially predicted through incongruence between single-gene and organismal phylogenies. Further examination of these predictions revealed that for a large proportion (56.4%) of these genes, homologous gene replacement (or independent gains) from outside Listeria (and consequent generation of COGs containing xenologous genes) was more probable than intra-Listeria transfers. In particular, only 104 (33%) of 322 high-confidence LGT predictions involved species from distinct Listeria clades. This observation is particularly striking in the light of the method used to infer LGT events. Phylogenetic incongruence from LGT will tend to be more pronounced when more distantly related organisms are involved and thus the method employed is expected to be more sensitive to such events. Rates of gene gain from outside Listeria were, by necessity, estimated in a distinct manner to intra-Listeria transfers, so it is not possible to make quantitative comparisons of intra- and intergenus transfer rates. High levels of conservation might render within-Listeria transfer events difficult to detect through phylogenetic approaches. However, the observation that individual core genes consistently yield well-resolved phylogenies leads us to speculate that gene exchange between Listeria subclades is relatively rare.
Finally, several striking examples of large-scale acquisition of functionally and genomically linked genetic loci were uncovered. Thirty-two genes involved in flagella synthesis (and the capacity for flagellar motility) were inferred, with high confidence, to have been gained in the common ancestor of L. grayi and Listeria sensu strictu. Phylogenetic reconstructions as well as gene content and arrangement emphasize the common descent of the listerial flagella with that present in the Bacillus cereus group, but not with those of other Bacilli and firmicutes. A simple explanation of the observed phylogenomic distribution of this flagella type would be that Listeria acquired their flagella from a B. cereus-like organism wherein (unlike in other Bacilli) the relevant genes are arranged in a single cluster. An alternative and perhaps equally valid scenario is that both B. cereus and Listeria acquired their current flagella from a similar, as yet unidentified, donor. We are aware of few reports of transfer of complete flagellar systems in bacteria (Liu and Ochman 2007; Poggio et al. 2007) and to our knowledge, this is the first report of such an event involving Gram-positive bacteria.
Although L. monocytogenes downregulates the expression of flagellar genes during pathogenesis through a temperature-dependent mechanism, several lines of evidence indicate that functional flagella augment the pathogenic potential of both B. cereus-like organisms (Bouillaut et al. 2005; Ramarao and Lereclus 2006) and Listeria (Dons et al. 2004). Accordingly, it is possible that the gain of flagella at least contributed to the evolution of pathogenicity in the ancestor of Listeria sensu strictu, whereas the presence of this organelle in L. grayi may contribute to its capacity to behave as a pathogen particularly in immune-compromised hosts (Rapose et al. 2008; Salimnia et al. 2010).
Another gain of linked genes occurred in a direct ancestor of Listeria sensu strictu, resulting in the formation of the propane-2-diol/ethanolamine/cobalamin locus, at least parts of which have been shown to be essential for pathogenesis of L. monocytogenes (Mellin et al. 2014) and other bacteria (Conner et al. 1998; Joseph et al. 2006; Klumpp and Fuchs 2007; Srikumar and Fuchs 2010). The genes from the eut operon are most closely related to homologs from other firmicutes, although consensus network representation of gene trees suggests potential recombination within the cluster. Although it was not possible to identify possible donors of the listerial eut cluster, genes from a homologous locus present in the genomes of Brocothrix species do not consistently emerge as sister to the listerial sequences, supporting the probabilistic inference of gene gain rather than differential loss of ancestral eut genes in basal Listeria.
Genes from the pdu operon most closely resemble homologs from Streptococcus, Pediococcus and Lactobacillus, but show high levels of conservation of sequence and arrangement with pdu operons from proteobacteria, whereas the cob–cbi cluster of Listeria most closely resembles homologous clusters from proteobacteria.
Although current genome sampling does not facilitate identification of donor genomes for any of these operons, it should be noted that, of available genomes, only in Listeria sensu strictu are pdu, eut and cob–cbi operons arranged as a single, tightly juxtaposed cluster, an intriguing observation given the dependence of both propane-2-diol and ethanolamine metabolism on the cobalamin cofactor produced by genes in the cob–cbi pathway.
Mechanisms possibly underlying the acquisition of both the flagellar and eut/pdu/cob–cbi clusters include transducing phages and integrative and conjugative elements (ICEs) (Wozniak and Waldor 2010). Recognition of specific cell-wall teichoic acid glycosylation modifications has recently been implicated in the wide host range specificity of transducing phages of Gram-positive bacteria and some such phages have been shown to be capable of transducing complex pathogenicity loci, for example, between Staphylococcus and Listeria (Winstel et al. 2013). We consider a contribution of phage-mediated transduction to transfer between Gram-positive bacteria to be very plausible. ICEs are mobile elements that mediate their own, sequence specific, integration and excision and which are transferred through conjugation. Listeria monocytogenes, and at least some Enterococcus, Bacillus, Staphylococcus and Streptococcus genomes contain related ICEs (Burrus et al. 2002) and ICEBs1 transfer from Bacillus subtilis into Bacillus anthracis, Bacillus licheniformis, and L. monocytogenes has been experimentally demonstrated (Auchtung et al. 2005). The L. monocytogenes ICELm1 element, the closest homolog of ICEBs1, is situated within 30 kb of the 5′-end of the pdu/eut/cob–cbi cluster. Furthermore, we note the presence of a broken integrase gene showing highest similarity to orfA of ICESt1 in the vicinity of the flagellar regulon of L. monocytogenes. ICELm1 is a part of the TN916-like family of elements, members of which have been identified in, among other taxa, Listeria, Enterococcus, Lactobacillus, Peptostreptococcus, Staphylococcus, Streptococcus, Veillonella, Fusobacterium, and proteobacteria (Roberts and Mullany 2009). Tn916 elements can be naturally transferred between Gram-negative and Gram-positive species (Bertram et al. 1991) and given the aforementioned considerations, may represent good candidates as mediators of the complex gene gain events inferred to have occurred within the genus Listeria.
Our findings are for the most part consistent with previous comparative genomic analyses of Listeria sensu strictu (Weller et al. 2014), wherein the core genome appears to be essentially closed with available sampling. Furthermore, our analyses indicate that lateral gene transfer, at least within Listeria, is not likely to be widespread. However, consideration of available genomes of basal Listeria species highlights distinct evolutionary dynamics where gene gain is more frequent and genome size apparently less constrained. For these organisms, available data support frequent gene gain, both by transfer from outside Listeria and by gene duplication. Strikingly, the distribution of inferred gene gain and loss events is consistent with recent studies suggesting that the rapid turnover of novel genetic material in individual prokaryotic genomes represents a taxonomically widespread phenomenon.
Supplementary Material
Supplementary methods and results, figures S1 and S2, text, and tables S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/ ).
Acknowledgments
This work was carried out thanks to the research infrastructures made available by the italian nodes of ELIXIR and LIFEWATCH (Molecular Biodiversity Lab) and supported by ELIXIR-EXCELERATE (H2020-INFRADEV-1-2015-1) and EMBRIC (H2020-INFRADEV-1-2014-1) projects as well as funding from the Italian Ministry of Health (IZSPB 006/10).
Literature Cited
- Abby SS, Tannier E, Gouy M, Daubin V. 2010. Detecting lateral gene transfers by statistical reconciliation of phylogenetic forests. BMC Bioinformatics 11:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A. 102:12554–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram J, Stratz M, Durre P. 1991. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J Bacteriol. 173:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch D, et al. 2012. Listeria fleischmannii sp. nov., isolated from cheese. Int J Syst Evol Microbiol. 63:526–532. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Bouillaut L, et al. 2005. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl Environ Microbiol. 71:8903–8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol Med Microbiol. 35:207–213. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cohen O, Ashkenazy H, Belinky F, Huchon D, Pupko T. 2010. GLOOME: gain loss mapping engine. Bioinformatics 26:2914–2915. [DOI] [PubMed] [Google Scholar]
- Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci U S A. 95:4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalquen DA, Altenhoff AM, Gonnet GH, Dessimoz C. 2013. The impact of gene duplication, insertion, deletion, lateral gene transfer and sequencing error on orthology inference: a simulation study. PLoS One 8:e56925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker HC, Bundrant BN, Fortes ED, Orsi RH, Wiedmann M. 2010. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl Environ Microbiol. 76:6085–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker HC, Cummings CA, et al. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker HC, et al. 2014. Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int J Syst Evol Microbiol. 64:1882–1889. [DOI] [PubMed] [Google Scholar]
- den Bakker HC, Manuel CS, Fortes ED, Wiedmann M, Nightingale KK. 2013. Genome sequencing identifies Listeria fleischmannii subsp. coloradonensis subsp. nov., isolated from a ranch. Int J Syst Evol Microbiol. 63:3257–3268. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bernal G, et al. 2006. A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mol Microbiol. 59:415–432. [DOI] [PubMed] [Google Scholar]
- Dons L, et al. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect Immun. 72:3237–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. 2010. A new pathway for the synthesis of alpha-ribazole-phosphate in Listeria innocua. Mol Microbiol. 77:1429–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 537:113–137. [DOI] [PubMed] [Google Scholar]
- Hain T, Steinweg C, Chakraborty T. 2006. Comparative and functional genomics of Listeria spp. J Biotechnol. 126:37–51. [DOI] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P. 2006. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 4:423–434. [DOI] [PubMed] [Google Scholar]
- Hansson M, Hederstedt L. 1992. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J Bacteriol. 174:8081–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson M, Rutberg L, Schroder I, Hederstedt L. 1991. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J Bacteriol. 173:2590-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- Huson DH, Dezulian T, Klopper T, Steel MA. 2004. Phylogenetic super-networks from partial trees. IEEE/ACM Trans Comput Biol Bioinform. 1:151-158. [DOI] [PubMed] [Google Scholar]
- Huson DH, Rupp R, Berry V, Gambette P, Paul C. 2009. Computing galled networks from real data. Bioinformatics 25:i85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, et al. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, et al. 2012. Population genomics of Chlamydia trachomatis: insights on drift, selection, recombination, and population structure. Mol Biol Evol. 29:3933-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153:1207-1220. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenne C, et al. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Halter E, Neuhaus K, Scherer S. 2013. Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int J Syst Evol Microbiol. 63:641-647. [DOI] [PubMed] [Google Scholar]
- Layer G, Reichelt J, Jahn D, Heinz DW. 2010. Structure and function of enzymes in heme biosynthesis. Protein Sci. 19:1137-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq A, et al. 2009. Listeria rocourtiae sp. nov. Int J Syst Evol Microbiol. 60:2210-2214. [DOI] [PubMed] [Google Scholar]
- Liu R, Ochman H. 2007. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci U S A. 104:7116-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, et al. 2013. A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc Natl Acad Sci U S A. 110:13132-13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, et al. 2014. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345:940-943. [DOI] [PubMed] [Google Scholar]
- Mishra KK, Mendonca M, Aroonnual A, Burkholder KM, Bhunia AK. 2011. Genetic organization and molecular characterization of secA2 locus in Listeria species. Gene 489:76-85. [DOI] [PubMed] [Google Scholar]
- Mistry J, Bateman A, Finn RD. 2007. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics 8:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville BA, et al. 2012. Characterization of pro-inflammatory flagellin proteins produced by Lactobacillus ruminis and related motile Lactobacilli. PLoS One 7:e40592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Wolf YI, Dubchak I, Koonin EV. 2009. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol. 191:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell RW, Green S, Laue BE, Sharp PM. 2014. The extent of genome flux and its role in the differentiation of bacterial lineages. Genome Biol Evol. 6:1514-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H. 2002. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet. 18:335-337. [DOI] [PubMed] [Google Scholar]
- Perkins JB, Pero JG. 2001. Vitamin biosynthesis. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its relatives: from genes to cells. Washington (DC): American Society for Microbiology; p. 279–293. [Google Scholar]
- Poggio S, et al. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J Bacteriol. 189:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Chakraborty T, Goebel W, Cossart P. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 60:1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarao N, Lereclus D. 2006. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 8:1483-1491. [DOI] [PubMed] [Google Scholar]
- Rapose A, Lick SD, Ismail N. 2008. Listeria grayi bacteremia in a heart transplant recipient. Transpl Infect Dis. 10:434-436. [DOI] [PubMed] [Google Scholar]
- Raux E, et al. 2003. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem J. 370:505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251-258. [DOI] [PubMed] [Google Scholar]
- Salimnia H, Patel D, Lephart PR, Fairfax MR, Chandrasekar PH. 2010. Listeria grayi: vancomycin-resistant, gram-positive rod causing bacteremia in a stem cell transplant recipient. Transpl Infect Dis. 12:526-528. [DOI] [PubMed] [Google Scholar]
- Schmid MW, et al. 2005. Evolutionary history of the genus Listeria and its virulence genes. Syst Appl Microbiol. 28:1-18. [DOI] [PubMed] [Google Scholar]
- Srikumar S, Fuchs TM. 2010. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl Environ Microbiol. 77:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Ling Y, Yu J, Wu J, Xiao J. 2013. Small proteins: untapped area of potential biological importance. Front Genet. 4:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, et al. 2014. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol. 6:2866-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, et al. 2008. A systematic survey of mini-proteins in bacteria and archaea. PLoS One 3:e4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AS, Archuleta J, Feng WC, Setubal JC. 2010. Missing genes in the annotation of prokaryotic genomes. BMC Bioinformatics 11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller D, Andrus A, Wiedmann M, den Bakker H. 2014. Listeria booriae sp. nov, and Listeria newyorkensis sp. nov., from food processing environments in the United States. Int J Syst Evol Microbiol. 65:286-292 [DOI] [PubMed] [Google Scholar]
- Welshimer HJ. 1963. Vitamin requirements of Listeria Monocytogenes. J Bacteriol. 85:1156-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshimer HJ, Meredith AL. 1971. Listeria murrayi sp. n.: a nitrate-reducing mannitol-fermenting Listeria. Int J Syst Bacteriol. 21:3-7. [Google Scholar]
- Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 18:691-699. [DOI] [PubMed] [Google Scholar]
- Winstel V, et al. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun. 4:2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 8:552-563. [DOI] [PubMed] [Google Scholar]
- Wurtzel O, et al. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 8:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.