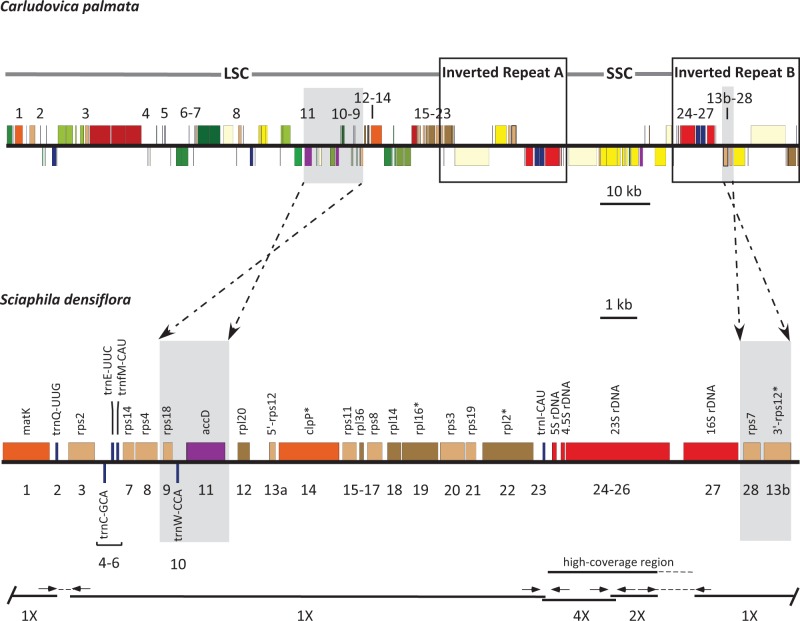
Fig. 3.—
Comparison of linearized plastomes of C. palmata (Cyclanthaceae) and S. densiflora (Triuridaceae). Boxes indicate IR regions (two copies, A and B) in Carludovica. Dashed arrows indicate predicted inversions of the small blocks highlighted in gray. Black lines below the Sciaphila plastome map indicate individual contigs (numbers below the lines indicate the estimated relative depth of coverage, see text). Gaps and contig overlap were, respectively, connected or confirmed using Sanger sequencing with primers at positions indicated with short arrows (primers not to scale; thin dashed lines are sequenced regions not represented in de novo contigs). A sector with higher read depth is indicated (the extent of higher-coverage is uncertain because this sector overlaps with a region produced using Sanger sequencing, indicated with a dotted line; 4×, five times coverage; 2×, two times coverage, see main text). Numbers indicate the 28 genes retained in Sciaphila, 18 of which are protein-coding (note that rps12 is a trans-spliced gene, noted here as 13a and 13b); *Genes with introns. The scale bars indicate relative plastome sizes of Carludovica and Sciaphila (kb, kilobase).