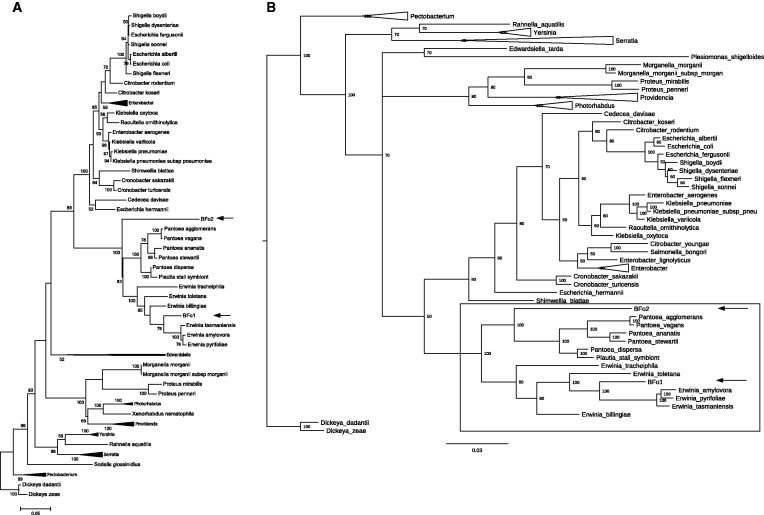
Fig. 1.—
Phylogenetic reconstruction showing placement of BFo bacteria among closely related Enterobacteriales. Phylogenetic tree constructed using a 5-protein MLSA. Gap-free alignment of five protein sequences giving a total of 833 positions. (A) ML consensus tree was constructed using the LG+G model with five categories and a +G parameter of 0.695. The initial tree for heuristic searching was obtained using the NJ algorithm. Bootstrap analysis with 1,000 pseudoreplicates was performed to infer nodal support. Bootstrap values below 70% not shown. Homogenous clades have been collapsed. Arrows indicate BFo. (B) 50% majority rule consensus BI phylogeny using the model=protein, Nst=1, Nbetecat=5, and default priors. Numbers at nodes indicate percentage node probably. Probabilities <50% not shown. Shaded area indicates the positions of the Pantoea and Erwinia genera (+both BFo bacteria). Homogenous clades have been collapsed. Arrows indicate BFo. Both trees are outgroup rooted along the branch leading to Dickeya.