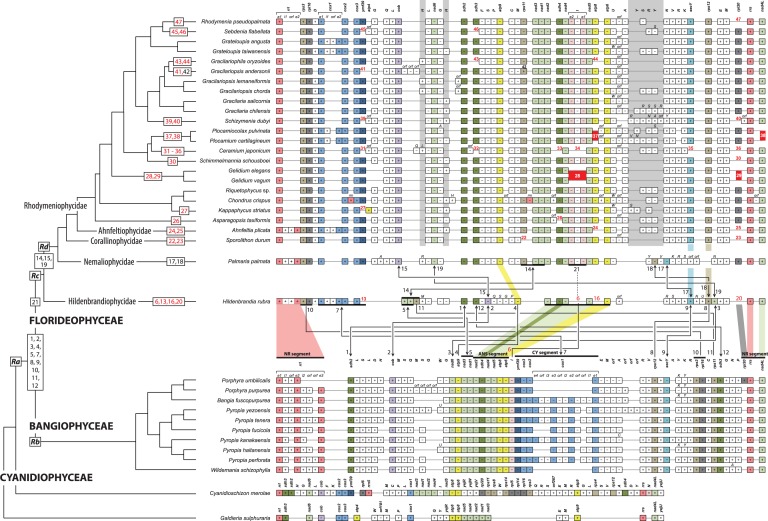
Fig. 2.—
Mitochondrial genome evolution in red algae. Mitochondrial gene order of 27 red algal species is illustrated along the best-supported phylogeny and associated taxonomy. Three classes (BANGIOPHYCEAE, CYANIDIOPHYCEAE, and FLORIDEOPHYCEAE) and five subclasses (Ahnfeltiophycidae, Corallinophycidae, Hildenbrandiophycidae, Nemaliophycidae, and Rhodymeniophycidae) of the red algal taxonomic system are indicated on the branches. Gene synteny is anchored by rrl (mt-rRNA large subunit) at the start and end rrs (mt-rRNA small subunit) at the end followed by nad4L, which shared an arrangement in all species (NR segment). Protein-coding and rRNA genes are abbreviated as shown in supplementary figure S1, Supplementary Material online. The tRNA genes are labeled using the one-letter code of their corresponding amino acid. The box indicates only gene existence, and box size does not correspond to gene size. The color code for genes corresponds to supplementary figure S1 and table S1, Supplementary Material online. Plus (+, forward) and minus (−, reverse) refer to the direction of gene transcription. The one letter code on the tRNA gene box indicates when alternative tRNA genes exist. Light-gray vertical boxes (three positions) point to variable tRNA gene positions in the Ahnfeltiophycidae, Corallinophycidae, and Rhodymeniophycidae. Evolutionary changes in the position of protein-coding genes are indicated with numbers (1–46) and mapped on the phylogeny; black numbers with blunt lines between genomes indicate gene rearrangement (e.g., no. 4); gene rearrangements with inversion are shown a line ended with arrowheads (e.g., no. 1); gene losses are designated with red numbers (e.g., no. 6); multigene rearrangements are marked with a thick horizontal line, that is, no. 5, 7, 10, and 14; and the trnI-intron region event (no. 21) is mapped on Palmaria and on the remaining phylogeny and corresponds to acquisition time. Strictly conserved regions are denoted with colored shadows between clades; that is, conserved ANS (nad2-sdh4-nad4-nad5-atp8-atp6), CY (ymf39-cox3-cox2-cox1), and NR (rpl20-rrs-nad4L-rrl) segments between Bangiophyceae and Florideophyceae, atp9 between Hildenbrandia and Palmaria, and rps12-secY between Hildenbrandia and Sporolithon.