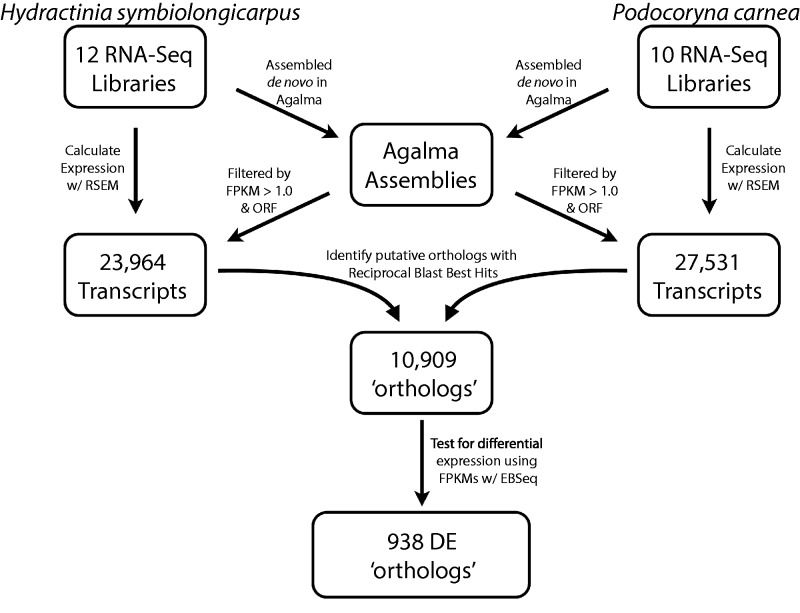
Fig. 2.—
Schematic of bioinformatics workflow. Initial transcriptomes for Hydractinia symbiolongicarpus and Podocoryna carnea are assembled from 12 and 10,100 bp paired-end Illumina libraries, respectively, using the program Agalma. Each transcriptome is filtered for transcripts that meet the Transdecoder reading frame criteria (as implemented in Agalma) and have an FPKM ≥ 1.0. Expression values are estimated for these remaining transcripts from each library independently using RSEM. Orthologs are identified using one-to-one reciprocal BLAST best hits between the transdecoder protein translations of the subsetted transcriptome using BLASTp under default setting. DE analyses are performed with EBSeq using FPKM and TPM expression normalizations.