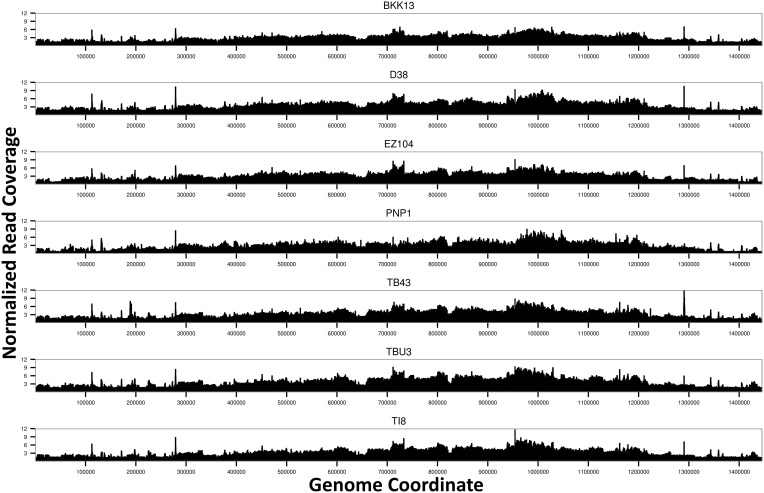
Fig. 3.—
Genome-wide read coverage for the seven wAnaITG genomes. Per-site read depth normalized against the D. ananassae nuclear genome (scaffold 13340) coverage is shown against the reference wRi genome coordinate. Note that regions of tRNA and rRNA, comprising 0.48% of the genome, were expected to have spurious reads from other endosymbiotic bacteria, leading to spikes of read coverage and were ignored from subsequent analysis (see text).