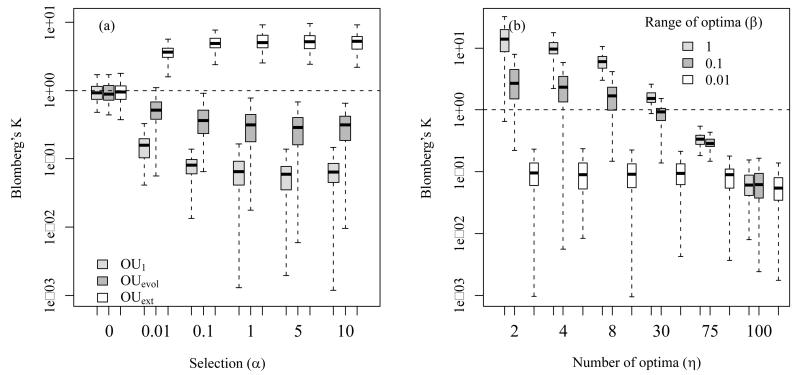
Figure 2.
We measured phylogenetic signal (PS, based on Blomberg’s K) in virtual data that we simulated with different niche evolution models. In the simulations niche values evolved along phylogenetic trees under three different niche evolution models: OU1 is a single optimum OU model where we set the optimum value equal to the root value; OUevol is a OU model where we set the optima of nodes to the niche values of the respective ancestor nodes; OUext is a OU model where η different clades differ strongly in their external selective optima (β indicates the range of different optima) and these optima also differ strongly from the root value (see Appendices S1 and S2 for more detail).
We found that, phylogenetic signal decreased with parameter changes assumed to increase PNC, such as high values of selection strength (α) in the OU1 and OUevol model. Phylogenetic signal, however, increased with parameter changes assumed to increase niche lability in the OUext model, such as moderate to high values of selection strength (α, panel a), high numbers of distinct selective optima (η, panel b) and increased ranges of selective optima niche values (high β, panel b). In all simulations, the variance of BM was set to σ=0.01. In plot a, the three models are parameterized as follows: η=1 for OU1, η=10 and β=1 for OUext and for OUevol the optima are equal to the ancestor nodes; in plot b, α=1. Each boxplot represents 100 repetitions. Note that y-axes are logarithmic and that outliers were not plotted.