Fig. 2.
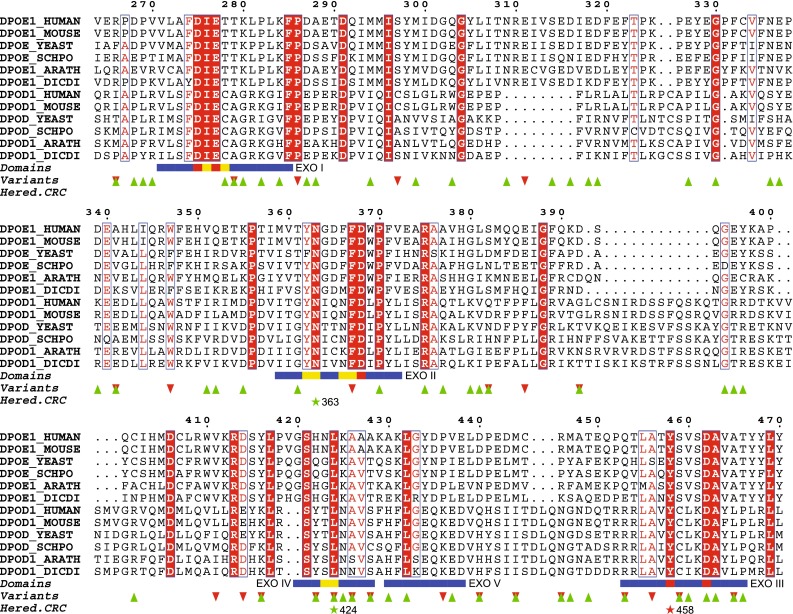
Multiple alignment of orthologous Polε and Polδ amino acid sequences. The alignment shows conserved positions in blue boxes (boxes with red background indicate completely conserved positions). The exonuclease domains (I–V) are indicated by horizontal blue lines. Essential residues of the DEDDy subfamily are indicated by yellow (active site residues) and red (catalytic residues) squares within the exonuclease domains. Known variants according to COSMIC and ExAC/dbSNP are indicated with red (filled triangle) and green (filled inverted triangle) triangles, respectively. The positions of the previously identified pathogenic germline mutations in CRC, p.Leu424Val and p.Asn363Lys, are indicated by green stars and sequence positions. The position of the variant identified in this study, p.Tyr458Phe, is indicated by a red star
