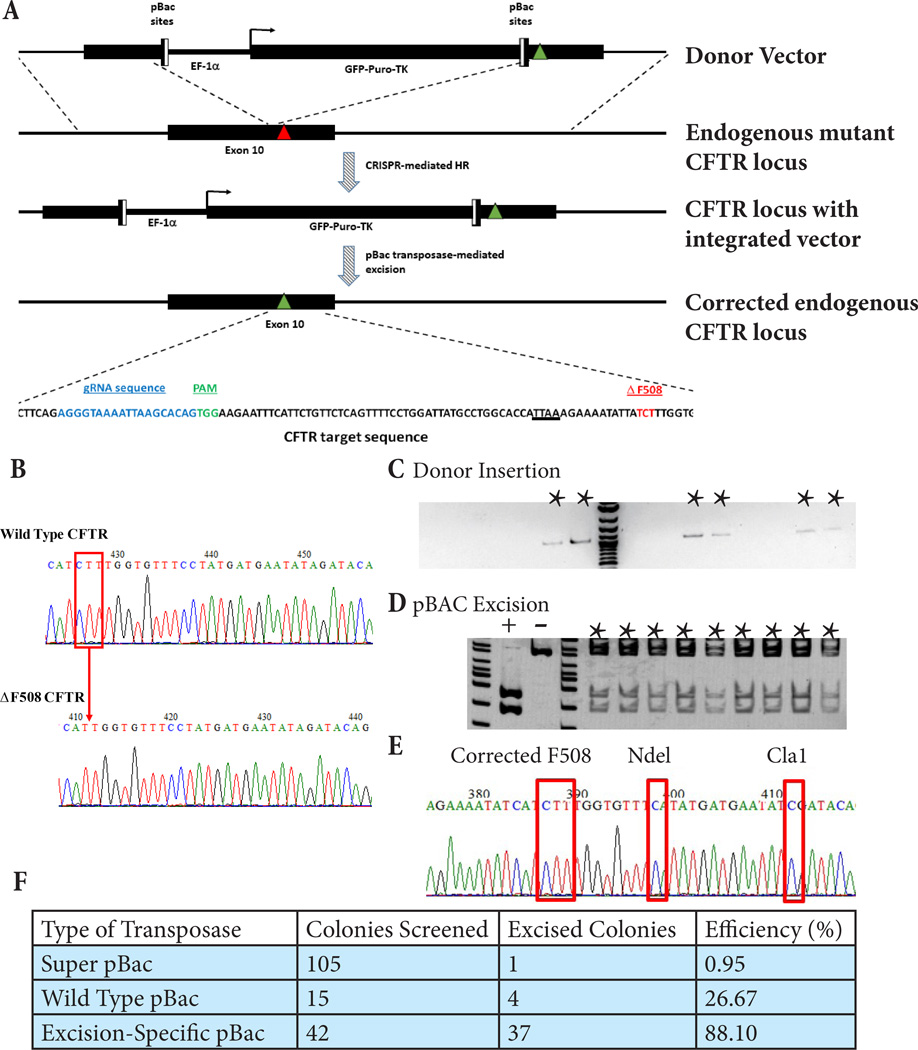
Figure 1. Footprint-free CRISPR-mediated correction of CF iPSC.
A) Schematic of strategy used for CRISPR-mediated correction of the CFTR ΔF508 mutation. Red triangle denotes the ΔF508 deletion (shown in red in the sequence at the bottom) and green triangles the correction of the deleted triplet of bases. The TTAA pBac recognition site is underlined. The actual CFTR target sequence is shown below with the gRNA target sequence in blue and the PAM in green. B) Sequencing analysis of the endogenous CFTR gene at the genomic locus of the ΔF508 mutation from wild type and CF patient-derived iPSC. The position of the CTT deletion in the mutant iPSC as compared to the wild type is indicated by the red box and arrow. C) Integration-specific PCR of puromycin resistant single-cell clones of CF iPSC after CRISPR treatment. Indicated clones contain the integrated selection cassette and corrective sequence in the correct position at the CFTR genomic locus. D) Screening by ClaI digest of a single-cell corrected clone after excision and negative selection of unexcised clones by ganciclovir. Indicated clones show precise and complete excision of the selection cassette from the endogenous CFTR genomic locus leaving behind only the corrected F508 sequence and the intended silent mutations encoding the restrictions sites used for screening here. E) Sequence analysis of corrected, excised clone showing the corrected ΔF508 mutation and adjacent silent mutations introduced in the endogenous CFTR gene. F) Table of excision screening done with different variations of the pBac transposase.