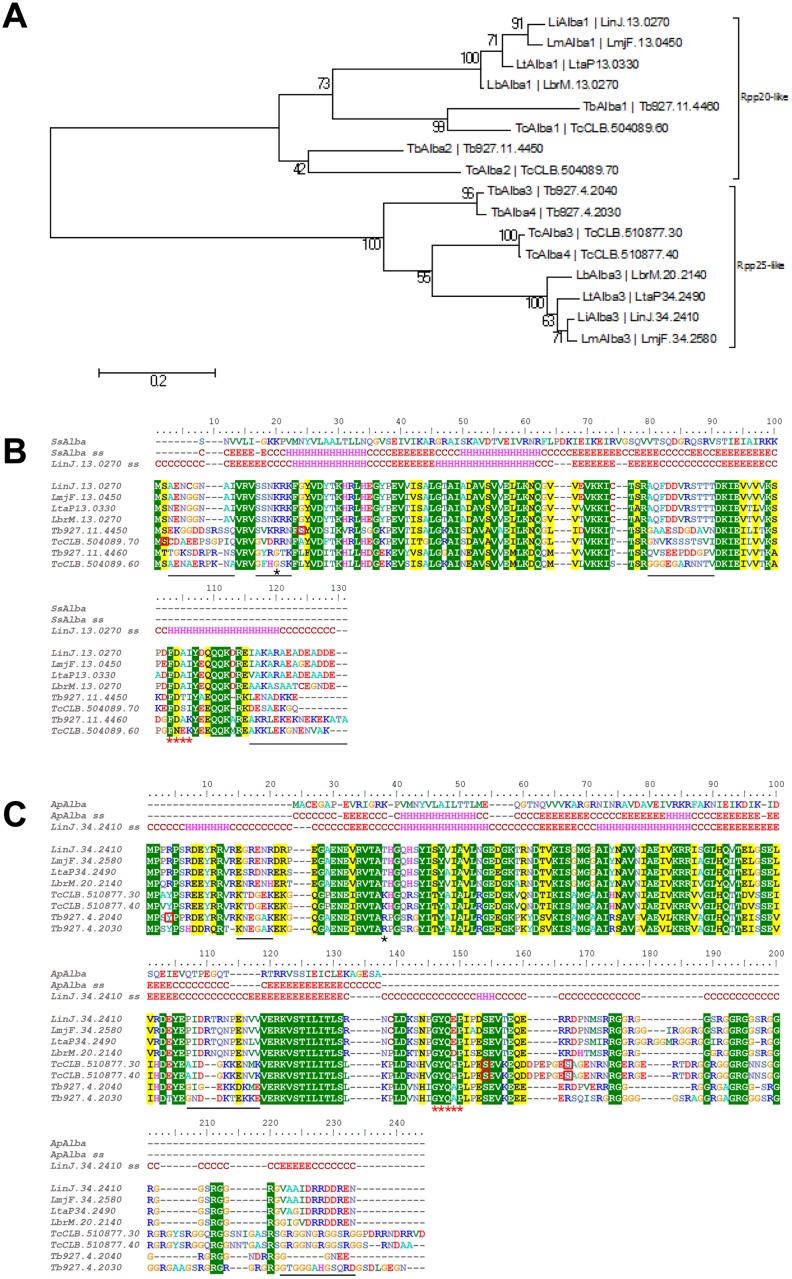
Fig 1. Sequence alignment and phylogeny of Alba-domain proteins in Leishmania, Trypanosoma brucei and T. cruzi species.
(A) Neighbor-joining tree showing the phylogenetic relationship between the Alba-domain proteins of TriTryps. Evolutional distances (scale) were estimated as the number of amino acid substitutions per site, considering Poisson correction. The two subgroups Rpp20-like and Rpp25-like are marked. (B) ClustalW alignment of Rpp20-like Alba-domain proteins merged with the in silico structure prediction of LiAlba1 (LinJ.13.0270) using the Phyre algorithm. The best score was obtained with the Alba protein from Sulfolobus solfataricus (Ss)(NCBI WP_010923153.1). ss: secondary structure in silico prediction; C: coil; H: helix; E: Sheet. Red squares indicate amino acids known to be phosphorylated on these specific genes (T. cruzi and T. brucei). The black star shows the expected position for Sir2 acetylation and the red stars underline the signature motif of the subgroup. Sequence variations in coiled regions between Trypanosoma spp. and Leishmania spp. are underlined with a black bar. LiAlba1 was used for Phyre structure prediction. (C) As in B for the Rpp25-like Alba-domain proteins. Structure prediction of LiAlba3 using Phyre outputs Alba2 from Aeropyrum pernix (Ap) K1 (NCBI WP_010866616.1) as the best match. LiAlba3 (LinJ.34.2410) was used for Phyre structure prediction. LinJ: L. infantum; LmjF: L. major; LtaP: L. tarentolae; LbrM: L. braziliensis; Tb: T. brucei; Tc: T. cruzi.