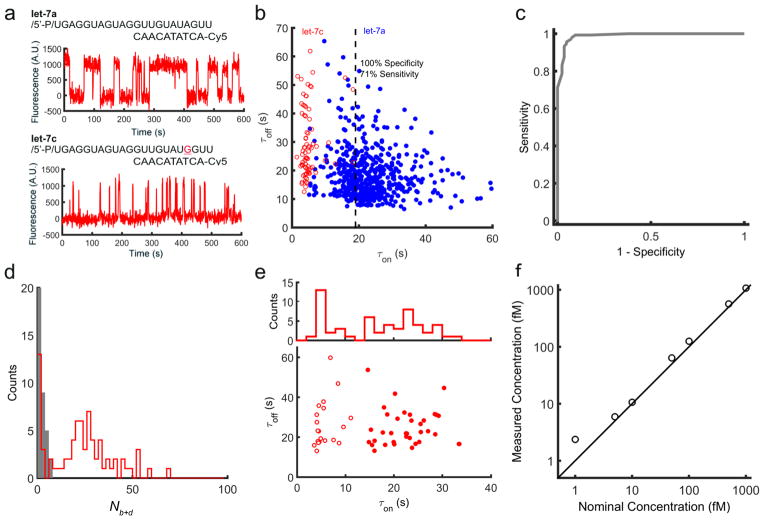
Figure 2. Single-molecule mismatch discrimination and detection of RNAs in crude biological matrices.
a, The fluorescent probe for let-7a exhibits long lifetimes of binding to let-7a (τon = 23.3 ± 8.3 s) but much more transient binding to let-7c (τon = 4.7 ± 3.0 s) due to a single mismatch. b, Dwell time analysis enables high-confidence single-copy-level discrimination between let-7a and let-7c. c, Receiver operating characteristic (ROC) plot constructed by varying the τon threshold for discriminating between let-7a and let-7c. d, Nb+d histogram for the detection of let-7 in HeLa cell extract in the presence or absence of the miRCURY let-7 inhibitor. e, Dwell times for molecules detected in HeLa extract using the fluorescent and capture probes for let-7a. The filled and open circles represent two clusters of target molecules classified by k-means clustering of τon values. f, Quantification of synthetic miR-141 spiked into human serum together with proteinase K and SDS.