Abstract
Entebbe bat virus (ENTV; Flaviviridae: Flavivirus), closely related to yellow fever virus, was first isolated from a little free-tailed bat (Chaerephon pumilus) in Uganda in 1957, but was not detected after that initial isolation. In 2011, we isolated ENTV from a little free-tailed bat captured from the attic of a house near where it had originally been found. Infectious virus was recovered from the spleen and lung, and the viral RNA was sequenced and compared with that of the original isolate. Across the polypeptide sequence, there were 76 amino acid substitutions, resulting in 97.8% identity at the amino acid level between the 1957 and 2011 isolates. Further study of this virus would provide valuable insights into the ecological and genetic factors governing the evolution and transmission of bat- and mosquito-borne flaviviruses.
Bat virus investigations in Uganda were initially inspired in the mid-1950s by the isolations of rabies virus and a novel flavivirus, later to become known as Rio Bravo virus (Flaviviridae: Flavivirus), from the salivary glands of insectivorous bats in the United States.1–3 Subsequently, investigators at the Uganda Virus Research Institute (UVRI) collected bats from the attic of UVRI, dissected salivary glands, and isolated viruses by intracerebral inoculation of triturated salivary gland extracts into neonatal mice.4,5 This first effort resulted in the isolation of Entebbe bat salivary gland virus, strain IL-30 (Flaviviridae: Flavivirus) from a little free-tailed bat (Chaerephon pumilus) (Cretzchmar).4,5 The taxon was then recognized by one of its synonyms: Tadarida (Chaerephon) limbata (Peters). Lumsden and others4,5 determined that this isolate was a novel flavivirus through a series of neutralization tests using immune sera against 20 arboviruses as well as against Rio Bravo virus. By 1964, bat virus research had expanded at UVRI, and sampling of 1,022 additional bats yielded 14 strains of viruses including Dakar bat virus (Flaviviridae: Flavivirus) also from little free-tailed bats, and Bukalasa bat virus (Flaviviridae: Flavivirus) from Angolan free-tailed bats (Mops condylurus) (A. Smith).6 Isolation of Mount Elgon bat virus (Rhabdoviridae: Ledantevirus) followed shortly thereafter from an eloquent horseshoe bat (Rhinolophus hildebrandtii eloquens) (K. Anderson) captured in Kenya, with the virus being isolated and characterized at UVRI.7,8 Additional bat virus discoveries in Uganda later included Kasokero virus (Bunyaviridae, unassigned) from an Egyptian fruit bat (Rousettus aegyptiacus) (E. Geoffroy).9 Entebbe bat virus (ENTV, renamed), Bukalasa bat virus, and Dakar bat virus were all isolated from insectivorous bats captured in the attic of UVRI.10 Serological surveys suggested that the infection prevalence of ENTV in wild populations of little free-tailed bats was high, but ENTV was never isolated again after the original discovery.10,11
To follow up on these early studies, arbovirus surveillance of bats was conducted in Uganda from 2011 to 2013. Bats were captured from attics in several locations around Entebbe and Kampala as part of a larger, country-wide sampling effort to be reported elsewhere. All bat captures were conducted under the approval of CDC/DVBD IACUC protocol 010-015. Bats were captured in attics using mist nets, taking appropriate biosafety precautions. On capture, bats were placed individually in cloth holding bags. Bats were anesthetized with halothane, exsanguinated by cardiac puncture, and euthanized by halothane overdose and cervical dislocation. Blood from bats was collected directly into serum separator tubes, centrifuged in the field, and placed immediately in a nitrogen dry vapor shipper. Liver, spleen, heart, lung, and kidney were all collected directly into cryovials and stored immediately on dry ice. In total, 95 little free-tailed bats and 34 Angolan free-tailed bats were captured in 2011, 15 little free-tailed bats in 2012, and 70 little free-tailed bats in 2013.
Virus isolation was attempted first from spleens, and on a positive result, virus isolation was also performed from the remaining tissues harvested from the infected bat. A single virus was isolated from 1 of 180 (0.5%) little free-tailed bats. The infected bat was an adult male, captured in Banga, Nakiwogo (0°04.884′ N, 32°27.030′ E) on June 23, 2011, near UVRI. Infectious virus was isolated from the spleen and lung; no infectious virus was recovered from the heart, liver, or kidney. No other tissues, oral or fecal swabs were collected from the insectivorous bats captured in 2011. The isolate was initially identified as ENTV by sequencing of the flavivirus NS5 amplicon using primers FU2/CFD3,12 followed by next-generation sequencing (NGS). Virus isolations, nucleic acid preparations, and NGS were performed as described previously.13 Novel primers were designed from contigs generated from the NGS data using Primer 3 software (Whitehead Institute for Biomedical Research, Cambridge, MA)14 (Supplemental Table 1), and the sequence for the entire genome was confirmed by direct sequencing. End terminal sequences were obtained using 5′/3′ RACE (FirstChoice® RLM-RACE, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Virus-specific RACE primers are provided in Supplemental Table 1. The 5′ outer reaction used primer 514R in combination with the 5′ RACE outer primer. The 5′ RACE inner reactions used primer 388R together with the 5′ RACE inner primer; amplification was also achieved with primers 15F + 388R. The 3′ outer reaction used either primer 9882F or 10328F in combination with the 3′ RACE outer primer. The 3′ inner RACE was not successful. A maximum likelihood phylogenetic tree was generated in MEGA version 6.015 (Center for Evolutionary Medicine and Informatics, Biodesign Institute, Tempe, AZ) using 10,493 nucleotides spanning the open reading frame sequence of select flavivirus genomes available in GenBank (Figure 1 ). The analysis was performed using a complete deletion substitution model and 1,000 bootstrap replicates.
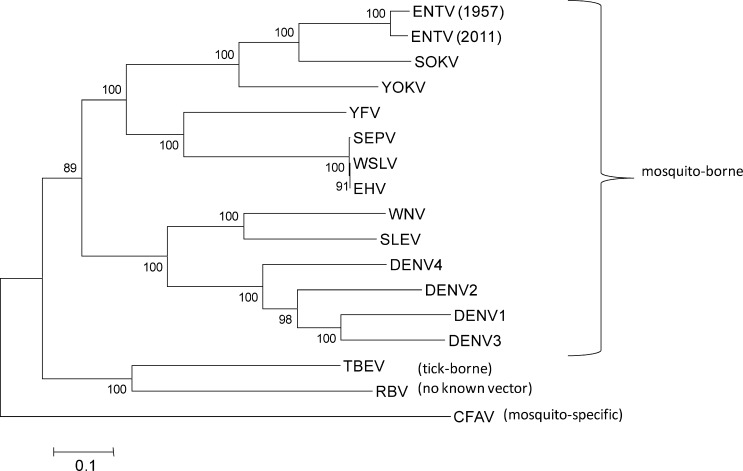
Figure 1.
Maximum likelihood tree based on open reading frame sequence (complete gap deletion). Scale bar indicates nucleotide substitutions per site. Bootstrap values > 50% are shown (1,000 replicates). CFAV = cell fusing agent virus (NC001564); DENV1 = dengue type 1 (M87512); DENV2 = dengue type 2 (M20558); DENV3 = dengue type 3 (M93130); DENV4 = dengue type 4 (AF326573); EHV = Edge Hill virus (DQ859060); ENTV = Entebbe bat virus (DQ837641); RBV = Rio Bravo virus (JQ582840); SEPV = Sepik virus (NC008719); SLEV = St. Louis encephalitis virus (NC007580); SOKV = Sokoluk virus (NC_026624); TBEV = tick-borne encephalitis virus (NC001672); WNV = West Nile virus (DQ211652); WSLV = Wesselsbron virus (DQ859058); YFV = yellow fever virus (K02749); YOKV = Yokose virus (NC005039); ZIKV = Zika virus (AF013415).
In total, 10,663 nucleotides of the ENTV genome were obtained (GenBank Accession number KP233893), of which the polypeptide is encoded between nucleotides 120 and 10,355. Kuno and Chang16 published the only other existing ENTV genome (DQ837641) sequenced from the original IL-30 strain. Compared with that original sequence, we report an additional 153 nucleotides in the 3′ noncoding region, but also experienced difficulty in obtaining the complete 3′ terminal sequence.16 Across the polypeptide sequence, there were 76 amino acid substitutions, resulting in 97.8% identity at the amino acid level between the 1957 and 2011 isolates. Before sequencing, the 1957 isolate had been passaged in suckling mouse brain, but the earlier passage history is unclear; the 2011 isolate was passaged twice in Vero cells. This 2.2% amino acid divergence is within the range that has been observed for other flaviviruses with isolations ranging between 1 and 49 years apart,17–19 indicating relatively little change in the ENTV genome in 54 years since the first isolation.
Every gene had at least one amino acid change except for PrM and M (Table 1). The highest percent divergence between the two isolates was in NS4A, 10.3% (Table 1). NS4A is a small, multifunctional, hydrophobic protein that plays a role in modulating the host's type I interferon response.20 It has also been shown to associate with host cellular vimentin during dengue virus replication to stabilize the replication complex.21 The functional significance of this high percentage of amino acid divergence has yet to be determined, but Hahn and others22 surmised that a large number of amino acid substitutions could be accommodated among the small, hydrophobic polypeptides NS2A, NS2B, NS4A, and NS4B so long as the hydrophobicity profile remained unchanged. In this case, changes to the NS4A hydrophobicity profile between these two isolates were inconsequential. Two of the 76 amino acid changes occurred at the junctions of NS2A/NS2B, and at NS4A/2K. The lengths of all of the genes were identical to those previously reported.16
Table 1.
Length of each gene region in the Entebbe bat virus genome and the number (%) of amino acid (aa) substitutions in the 2011 isolate (KP233893) relative to the 1957 isolate (DQ837641)
| Gene | Length (aa) | Number of aa substitutions (%) |
|---|---|---|
| C | 119 | 6 (5.0) |
| PrM | 93 | 0 (0.0) |
| M | 75 | 0 (0.0) |
| E | 489 | 5 (1.0) |
| NS1 | 353 | 11 (3.1) |
| NS2A | 228 | 9 (3.9) |
| NS2B | 130 | 1 (0.8) |
| NS3 | 620 | 7 (1.1) |
| NS4A | 126 | 13 (10.3) |
| 2K | 23 | 1 (4.3) |
| NS4B | 249 | 6 (2.4) |
| NS5 | 906 | 17 (1.9) |
The phylogenetic relationship between ENTV and yellow fever virus (YFV) is very interesting, as YFV is historically one of the most medically important mosquito-borne flaviviruses. Although ENTV falls phylogenetically within the mosquito-borne flaviviruses and historically has been placed in the YFV group,16,23 it has only been isolated twice from bats (1957 and 2011) and never from wild-caught mosquitoes or humans. Figure 1 confirms the phylogenetic placement of both isolates of ENTV, as published previously,12,16,23 and also supports the more recent International Committee on Taxonomy of Viruses placement of ENTV in the Entebbe bat virus group, a sister clade to the YFV group.24 Consistent with this new grouping, ENTV shared the closest sequence homology with Sokoluk virus, which has been isolated from insectivorous bats and soft ticks in Kyrgyzstan.25 Pathogenicity experiments were not conducted as part of this study, but originally ENTV was shown to be pathogenic to adult mice by the intracerebral route and for infant mice by both intracerebral and intraperitoneal routes.4,5 In our experience, it caused cytopathic effects and produced plaques on Vero cells. ENTV also replicates in mosquito cells.26 Despite remaining an obscure bat-associated virus for over a half century, ENTV continues to circulate among little free-tailed bats in Uganda. Further study of this virus would provide valuable insights into the ecological and genetic factors governing the evolution and transmission of bat-borne and mosquito-borne flaviviruses.
Supplementary Material
ACKNOWLEDGMENTS
We especially thank the homeowners who allowed us to capture bats in their homes for this study, David Ssekatawa and Godfrey Kyazze for transportation and technical assistance, and Jeff Borchert for helping with local arrangements. Bat collections in 2012 and 2013 were also assisted by Betty Nalikka, Nick Panella, Julian Kerbis, Luke Nyakarahuka, Eric Mossel, and Amy Gilbert.
Disclaimer: The findings and conclusions in this report are those of the authors only, and do not necessarily reflect the views of their respective institutions.
Footnotes
Financial support: This work was funded by an interagency agreement between the Centers for Disease Control and the United States Agency for International Development.
Authors' addresses: Rebekah C. Kading, Genesis Laboratories, Inc., Wellington, CO, E-mail: rcmosquito@gmail.com. Robert Kityo, Department of Biological Sciences, Makerere University, Kampala, Uganda, E-mail: kityrob@gmail.com. Teddie Nakayiki and Julius Lutwama, Department of Arbovirology, Uganda Virus Research Institute, Entebbe, Uganda, E-mails: nakayikiteddie@yahoo.com and jjlutwama03@yahoo.com. Jeremy Ledermann, Mary B. Crabtree, and Barry R. Miller, Division of Vector-Borne Diseases, CDC, Fort Collins, CO, E-mails: bpj7@cdc.gov, meb3@cdc.gov, and brm4@cdc.gov.
References
- 1.Johnson HN. Rabies. In: Thomas Francis., editor. Diagnostic Procedures for Virus and Rickettsial Diseases. 2nd edition. New York, NY: American Public Health Association; 1956. [Google Scholar]
- 2.Johnson HN. Proceedings of the 9th Pacific Science Congress. Vol. 17. Bangkok, Thailand: Pacific Science Association; 1962. The Rio Bravo virus: virus identified with group B arthropod-borne viruses by haemagglutination inhibition and complement fixation tests; p. 39. Abstract. [Google Scholar]
- 3.Burns KF, Farinacci CJ. Virus of bats antigenically related to St. Louis encephalitis. Science. 1956;123:227–228. doi: 10.1126/science.123.3189.227. [DOI] [PubMed] [Google Scholar]
- 4.Lumsden WHR, Williams MC, Mason PJ. A virus from insectivorous bats in Uganda. Ann Trop Med Parasitol. 1961;55:389. doi: 10.1080/00034983.1961.11686063. [DOI] [PubMed] [Google Scholar]
- 5.Lumsden WHR, Williams MC, Mason PJ. East African Virus Research Institute Report, July 1956–June 1957. Nairobi, Kenya: Government Printer; 1957. From bat salivary glands; p. 22. Publication of the East African High Commission. [Google Scholar]
- 6.Williams MC, Simpson DIH, Shepherd RC, O'Sullivan JP, Cunningham JC, Lule M. East African Virus Research Institute Report, July 1963–December 1964. Nairobi, Kenya: Government Printer; 1964. Virus isolations from bats; p. 42. Publication of the East African High Commission. [Google Scholar]
- 7.Metselaar D. No. 224 Mount Elgon bat strain BP846. Am J Trop Med Hyg. 1970;19:1119–1120. [PubMed] [Google Scholar]
- 8.Blasdell KR, Guzman H, Widen SG, Firth C, Wood TG, Holmes EC, Tesh RB, Vasilakis N, Walker PJ. Ledantevirus: a proposed new genus in the Rhabdoviridae has a strong ecological association with bats. Am J Trop Med Hyg. 2015;92:405–410. doi: 10.4269/ajtmh.14-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalunda M, Mukwaya LJ, Mukuye A, Lulu M, Sekyalo E, Wright J, Casals J. Kasokero virus: a new human pathogen from bats (Rousettus aegyptiacus) in Uganda. Am J Trop Med Hyg. 1986;35:387–392. doi: 10.4269/ajtmh.1986.35.387. [DOI] [PubMed] [Google Scholar]
- 10.Simpson DIH, Williams MC, O'Sullivan JP, Cunningham JC, Mutere FA. Studies on arboviruses and bats (Chiroptera) in east Africa II. Isolation and haemagglutination-inhibition studies on bats collected in Kenya and throughout Uganda. Ann Trop Med Parasitol. 1968;62:432–440. doi: 10.1080/00034983.1968.11686580. [DOI] [PubMed] [Google Scholar]
- 11.Shephard RC, Williams MC. Studies on viruses in east African bats (Chiroptera) Zoonoses Res. 1964;3:125–139. [PubMed] [Google Scholar]
- 12.Kuno G, Chang G-JJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kading RC, Gilbert AT, Mossel EC, Crabtree MB, Kuzmin IV, Niezgoda M, Agwanda B, Markotter W, Weil MR, Rupprecht CE, Montgomery J, Miller BR. Isolation and molecular characterization of Fikirini rhabdovirus, a novel virus from a Kenya bat. J Gen Virol. 2013;94:2393–2398. doi: 10.1099/vir.0.053983-0. [DOI] [PubMed] [Google Scholar]
- 14.Rozen S, Skaletsky HJ. Primer3. 1998. http://biotools.umassmed.edu/bioapps/primer3_www.cgi Available at. Accessed June 11, 2015. [DOI] [PubMed]
- 15.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolution Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuno G, Chang G-JJ. Characterization of Sepik and Entebbe bat viruses closely related to yellow fever virus. Am J Trop Med Hyg. 2006;75:1165–1170. [PubMed] [Google Scholar]
- 17.Faria NR, Nogueria RM, deFilippis AM, Simoes JB, Nogueria Fde B, DaRocha Queiroz Lima M, dos Santos FB. Twenty years of DENV-2 activity in Brazil: molecular characterization and phylogeny of strains isolated from 1990 to 2010. PLoS Negl Trop Dis. 2013;7:e2095. doi: 10.1371/journal.pntd.0002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu LJ, Liu R, Ye S, Ling H, Zhu CM. Genomic analysis of a newly isolated of Japanese encephalitis virus strain, CQ11-66, from a pediatric patient in China. Virol J. 2013;10:101. doi: 10.1186/1743-422X-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar SR, Patil JA, Cecilia D, Cherian SS, Barde PV, Walimbe AM, Yadav PD, Yergolkar PN, Shah PS, Padbidri VS, Mishra AC, Mourya DT. Evolution, dispersal and replacement of American genotype dengue type 2 viruses in India (1956–2005): selection pressure and molecular clock analyses. J Gen Virol. 2010;91:707–721. doi: 10.1099/vir.0.017954-0. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci USA. 2003;24:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teo CSH, Chu JJH. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J Virol. 2014;88:1897–1912. doi: 10.1128/JVI.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn CS, Dalrymple JM, Strauss JH, Rice CM. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc Natl Acad Sci USA. 1987;84:2019–2023. doi: 10.1073/pnas.84.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macdonald J, Poidinger M, Mackenzie JS, Russell RC, Doggett S, Broom AK, Phillips D, Potamski J, Gard G, Whelan P, Weir R, Young PR, Gendle D, Maher S, Barnard RT, Hall RA. Molecular phylogeny of Edge Hill virus supports its position in the yellow fever virus group and identifies a new genetic variant. Evol Bioinform. 2010;6:91–96. doi: 10.4137/ebo.s4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Amsterdam, The Netherlands: Elsevier Academic Press; 2012. IXth Report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- 25.L'vov DK, Al'khovski SV, Shchelkanov MIu, Shchetinin AM, Deriabin PG, Gitel'man AK, Samokhvalov EI, Botikov AG. Taxonomy of the Sokuluk virus (SOKV) (Flaviviridae, Flavivirus, Entebbe bat virus group) isolated from bats (Vespertilio pipistrellus Schreber, 1774), ticks (Argasidae Koch, 1844), and birds in Kyrgyzstan. Vopr Virusol. 2014;59:30–34. [PubMed] [Google Scholar]
- 26.Varelas-Wesley I, Calisher CH. Antigenic relationships of flaviviruses with undetermined arthropod-borne status. Am J Trop Med Hyg. 1982;31:1274–1284. doi: 10.4269/ajtmh.1982.31.1273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.