Fig. 5.
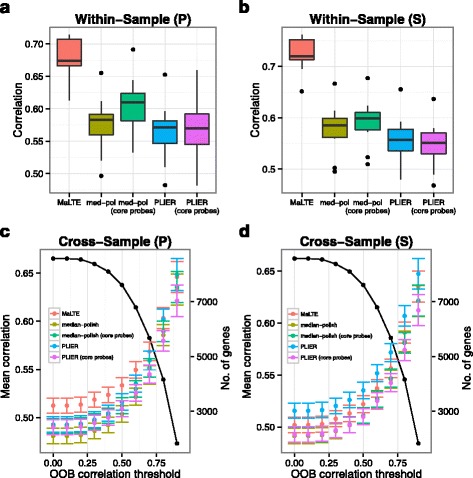
Application to archived data. MaLTE, trained using the GTEx data, was applied to predict gene expression from published microarray data based on brain samples for which RNA-Seq data was also available. Despite the fact that the two studies used different array platforms (Affymetrix Human Exon 1.0 ST arrays and Affymetrix Human Gene 1.1 ST arrays for the brain and GTEx studies, respectively), MaLTE predictions exceeded the within-sample correlations obtained using median-polish and PLIER. MaLTE predictions were based on probes shared between the two array platforms. Box plots of (a) Pearson and (b) Spearman within correlations are shown. (c) Pearson and (d) Spearman cross sample correlations with OOB filtering. The black line represents the number of genes/transcripts at each level
