Background
Gene expression silencing at mRNA level by microRNAs (miRNA) is a well-established form of post-transcriptional regulation1, 2. Such silencing is achieved through miRNA binding to miRNA response elements (MRE) residing mainly in the 3’ untranslated regions (UTRs) of the target mRNA. Over 1000 human miRNAs have been identified3, and the prevalence of miRNA regulation in a broad range of biological processes and disease often attributes to the fact that a single miRNA can repress hundreds of different mRNAs4. Interestingly, a single target mRNA often possesses MREs of distinct miRNAs in its 3’UTR5. Questions have been raised regarding the need for this redundancy in regulation and these multiple MREs were once thought to serve as regulatory buffers of different miRNAs. In a recent seminal work, a novel theory, termed the competing endogenous RNA (ceRNA), was proposed to provide a plausible explanation for this interesting phenomenon from a new perspective of gene regulation6. According to the ceRNA theory, MREs function as “letters” of this new regulatory system, and ceRNAs, or sets of RNAs including mRNA, pseudogenes, and long noncoding RNAs, can communicate, or regulate each other, through competition for common MREs. As such, ceRNA regulatory networks provide a unifying system for regulations among transcriptome-wide RNAs, greatly expanding the functions of RNAs6. Alteration of this competition between ceRNAs could modify normal state gene expression and in return alter the status of biological pathways to promote an oncogenic program for example. To that end, a PTEN ceRNA network was uncovered and shown to potentially regulate oncogenesis6.
The fact that this new level of RNA regulations could be prevalent in cells has prompted research to identify ceRNAs of genes related to disease. However, the complexity of ceRNA regulations and an incomplete knowledge of miRNA binding have hampered the prediction of ceRNAs, which often requires the use of computational tools and databases that are not readily available to the users. Thus far, two algorithms for human ceRNA predictions have been proposed. MuTaMe, proposed in6, aims to predict ceRNAs of a GOI. It starts by selecting a set of, ideally experimentally validated, miRNAs that target the given GOI in its 3’ UTR region. Predicted ceRNAs by sequence-pairing are the mRNAs that are also targeted by these miRNAs and the prediction is made based on scores generated from binding affinity statistics. While MuTaMe succeeded in predicting several ceRNAs of PTEN, it is not accessible for predicting other GOIs because experimentally validated miRNAs targeting a new GOI are mostly unavailable and binding affinity statistics used in MuTaMe are insufficient for accurate predictions. Furthermore, MuTaMe has not been implemented as a software tool yet and cannot be accessed by the general public. The second algorithm, Hermes, proposed in7, infers ceRNAs from expression profiles of genes and miRNAs by using conditional mutual information (CMI). While Hermes combines ceRNA/miRNA/target triplets via tissue specific gene expression, however, it does not provide an implementation that combines sequence-binding statistics with gene expression. There is a shortage of user-friendly tools that can be easily used for anyone interested in ceRNA research.
To address the need for user-friendly tools, we developed here TraceRNA, a web-based application for transcriptome-wide ceRNA discovery. TraceRNA is flexible, powerful, and user-friendly. It includes MiRTarBase8, a database of experimentally validated miRNA-target pairs, and miRNA binding scores and related data (sites position, length, etc.) from three prediction algorithms (SVMicrO9, BCMicrO10, and SiteTest) with different emphasis. TraceRNA provides the user with the flexibility to perform ceRNA predictions using one of three algorithms to meet different study objectives. Currently, TraceRNA maintains a database that includes genome-wide targets of >700 human miRNAs predicted by three algorithms. The user can compare among the prediction results from these different algorithms to either complement or reach a consensual prediction.
Two important observations have been integrated into the TraceRNA for context-specific ceRNA discovery. The first is that the miRNA expression is condition-specific. That is, if a miRNA is not expressed in a tissue environment or disease state, one can ignore its target-binding specificity. The second is that, GOI and its ceRNAs’ expressions are positively correlated because of the competition for miRNA binding. Therefore, an increased/decreased GOI expression will attract more/less miRNA binding away its ceRNAs, resulting in increased/decreased ceRNA expression level due to the decreased/increased repression effect of miRNAs. As another unique feature, TraceRNA can construct ceRNA interaction networks to help delineate complex interactions of ceRNAs and gain further insight into this novel ceRNA regulation-modulation mechanism. Finally, TraceRNA is developed to be user-friendly web application with an accessible interface. It generates predictions including statistics such as p-values and false discovery rate (FDR) both online and in spreadsheets available for download.
Methods
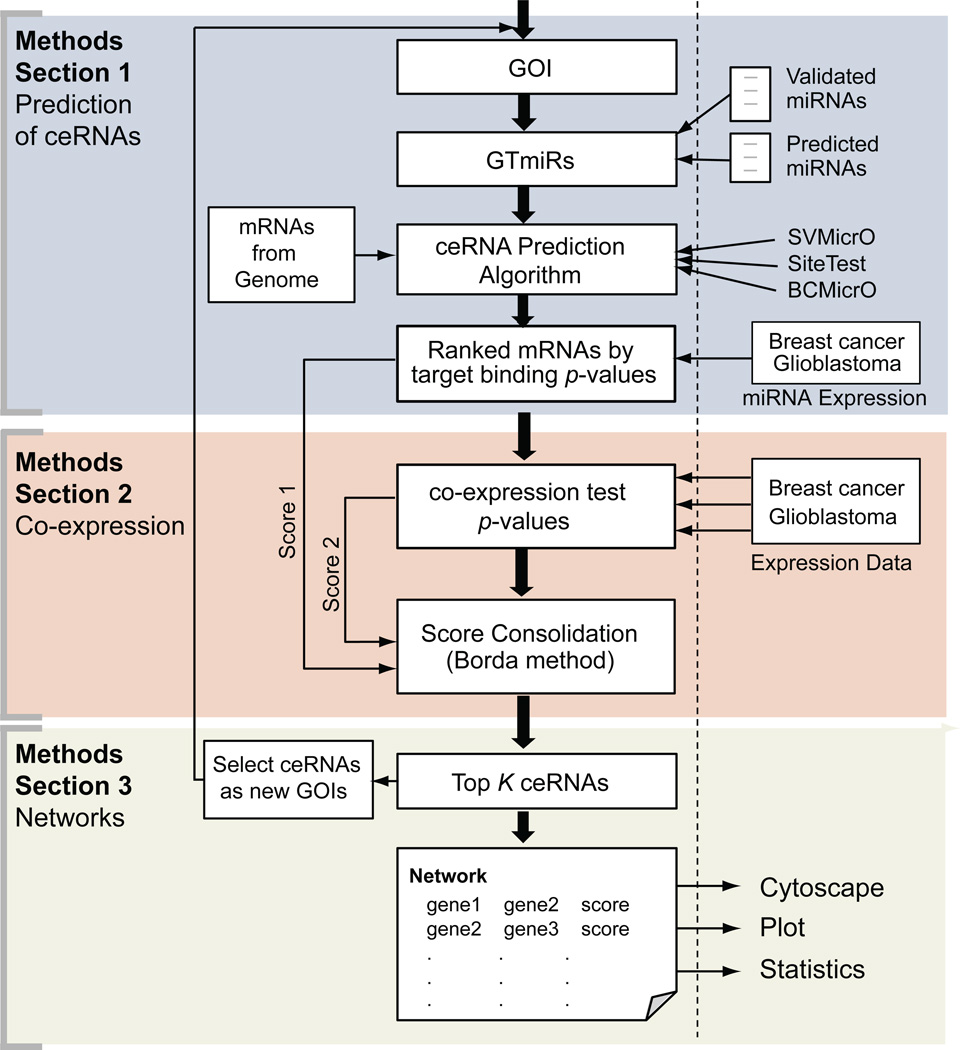
The goal of TraceRNA is to predict ceRNAs of a GOI, which are mRNAs that share MREs from a set of miRNAs that also target the GOI. In this paper, we named these miRNAs as GOI targeting miRNAs, or GTmiRs. ceRNAs’ competition for GTmiRs binding to GOI will alter the expression of GOI and its ceRNAs in a coordinated fashion and co-expression can be observed, where expressions of GOI and its ceRNAs are expected to be correlated. Predictions of ceRNAs can be done by examining miRNA:mRNA sequence pairing and/or GOI-ceRNAs co-expression. TraceRNA includes three main processing sections in its pipeline (Fig. 1): 1) sequence-based prediction of ceRNAs, 2) co-expression analysis of GOI and ceRNAs’ expression levels, and 3) generation of ceRNA regulatory network. Additionally, miRNA expression data is also included in TraceRNA for the user to select context-specific GTmiRs (Supplemental Fig. S1).
Figure 1.
TraceRNA Pipeline. The user initiates TraceRNA predictions with a gene of interest (GOI). Experimentally validated miRNAs and/or predicted miRNAs that target this GOI can be selected as GTmiRs. These GTmiRs are then fed to one of the three sequence-level target prediction algorithms (SVMicrO, SiteTest or BCmicrO) to generate a list of predicted ceRNAs by sequence-pairing together with the p-values and FDRs (Section 1). In addition, the user can select one of the provided expression sets (GMB and Breast Cancer datasets) to evaluate the expression correlation between the predicted ceRNAs by sequence-pairing and the GOI under the specific tissue/tumor condition and obtain predicted ceRNAs by co-expression test. Multiple prediction scores are consolidated with Borda method (Section 2). To generate a ceRNA network, top 20 ceRNAs will be selected as a set of the new GOIs, or cGOIs, each then subject to a round of new predictions to obtain their corresponding ceRNAs or cGOI-ceRNAs pairs. All resulting GOI-ceRNAs and cGOI-ceRNAs pairs with their scores are used to generate a ceRNA mediated regulatory network (Section 3).
Sequence-based prediction of ceRNAs
Selection of GOI targeting miRNAs (GTmiRs)
Given a GOI provided by the user, the first step in TraceRNA is to identify GTmiRs, or miRNAs that target GOI. TraceRNA provides two alternatives for GTmiRs identification (Fig. 1). First, TraceRNA maintains a local copy of experimentally validated miRNAs: targets pairs curated by miRTarBase Release 2.5 (downloaded on July 2012). Second, genome-wide SVMicrO9 predictions for >700 miRNAs were pre-calculated.. SVMicrO9 was developed previously to predict miRNA targets. It uses a support vector machine with sequence-based features including binding secondary structure, energy, binding conservation, number of predicted sites, and site densities. SVMicrO was tested to achieve improved performance compared to several popular algorithms including TargetScan, miRanda, Pictar, etc. The predicted miRNAs are displayed to the user in descending order of p-values (See Section 1.3). The user may select a subset or all of the miRNAs from these two sources as GTmiRs.
Prediction of ceRNAs
Once GTmiRs are selected, TraceRNA predicts ceRNAs as the mRNAs that are also targeted by these GTmiRs, by using one of three miRNA target prediction algorithms: 1) SVMicrO9, 2) BCMicrO10, or 3) SiteTest, depending on the user’s selection. SVMicrO9 and BCMicrO10 are two in-house developed algorithms which were published previously. As discussed above, SVMicrO makes predictions by utilizing a large number of miRNA binding features. BCMicrO uses a Bayesian approach that integrates prediction scores from 6 popular algorithms: TargetScan11, miRanda12, PicTar13, mirTarget214, PITA15, and DIANA micro-T16. Both algorithms provide more accurate predictions than existing algorithms. The prediction scores of SVMicrO and BCMicrO were pre-calculated and stored in a MySQL database. In addition, a new algorithm, SiteTest, inspired by MuTaMe6, was also developed and its pseudo code is included in Supplemental Materials.
In order to show the scores calculation, let Si be the score of GTmiR i targeting an mRNA by either algorithms and K be the total number of GTmiRs. Then, the score, S, for the mRNA to be a ceRNA predicted by sequence-pairing is calculated as
| (1) |
We discuss next the calculation of the predictions significance.
Statistical Significance of Predicted ceRNAs
We first discuss the calculation of statistical significance for the SVMicrO scores. According to (1), S is calculated as the average of the sequence-pairing scores of each GTmiR and the mRNA. To calculate the p-value for S, the distribution of S under the null hypothesis, i.e., the mRNA is not predicted by sequence-pairing as a ceRNA, needs to be obtained. Because S is the average Si, then the distribution of Si under the null hypothesis needs to be evaluated first. Adopting the method developed in BCMicrO, the empirical distribution Si under the null hypothesis was observed as a mixture of two distributions, one clustered around smaller scores and the other around larger scores (Fig. 2). Given that most genes are not miRNA targets and they should have smaller SVMicrO scores, we hypothesized that the distribution around smaller scores characterizes the scores derived from genes not targeted by any miRNA, which was further assumed to follow the i.i.d. Gamma distribution, or Si~ Gamma(α,β) whose parameters α and β were obtained from fitting the empirical scores Si (Supplemental Figure S1). Subsequently, due to (1), S is also Gamma-distributed under the null hypothesis as:
| (2) |
Therefore, the probability (p-value) of a sequence-pairing prediction score S can be evaluated analytically by (2). The same method was applied to BCMicrO and SiteTest by fitting the Gamma distributions directly to their scores. Once p-values of all predicted ceRNAs by sequence-pairing are calculated, the corresponding False Discovery Rates (FDRs) are computed using the Benjamini-Hochberg method17.
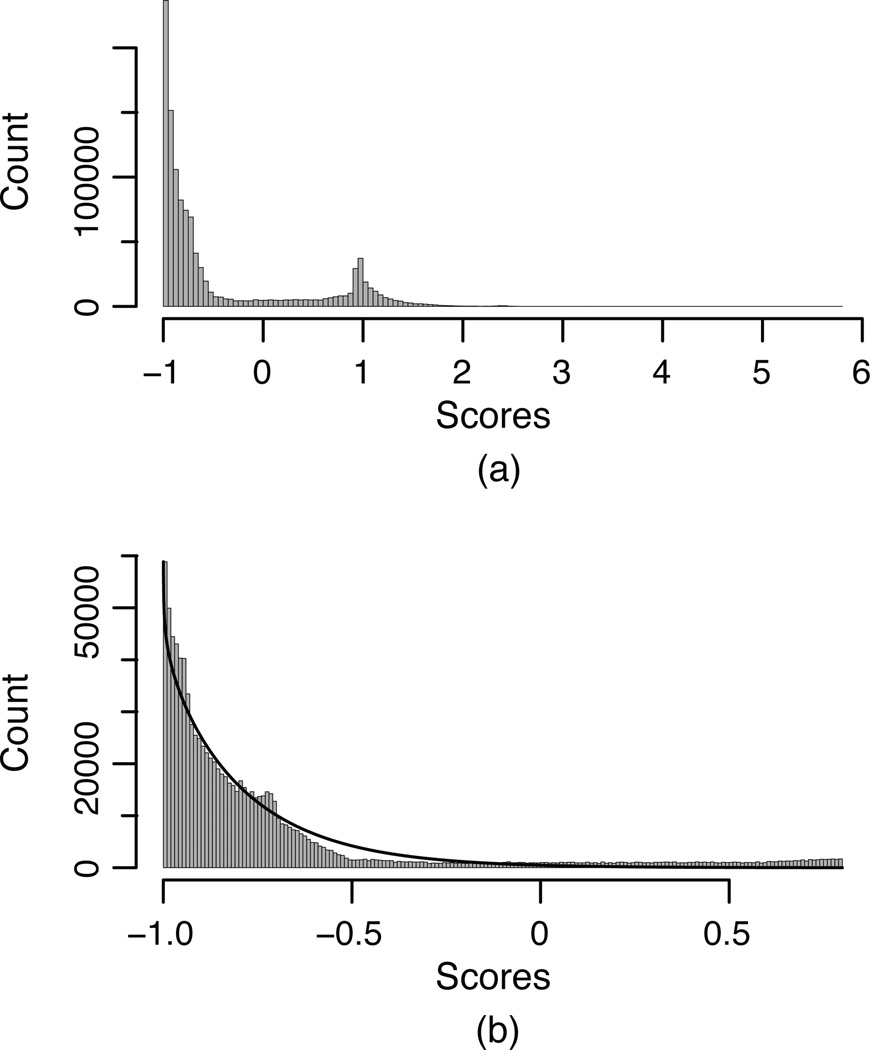
Figure 2.
Illustration of p-value calculation for ceRNA prediction scores. (a) Histogram of SVMicrO scores for genome-wide targets of 500 miRNAs. (b) Zoom-in view of the histogram in (a) and the fitted (shifted) Gamma distribution (solid line). The parameters of the fitted Gamma distribution are α=0.7234 and β=0.3594.
Co-expression based prediction of ceRNAs
Test for co-expression between GOI and predicted ceRNAs by sequence-pairing
TraceRNA can also integrate a tissue or disease specific expression dataset to predict tissue or disease specific ceRNAs of the GOI and potentially further improve the prediction specificity (Fig. 1). Currently, expression datasets of glioblastoma multiforme (GBM)18 and Breast Cancer19 from TCGA (http://cancergenome.nih.gov/) are included. The users may contact the web-master to upload their own expression datasets if needed. Because higher GOI expression competitively attracts more miRNA binding and thus reduces the possibility of the same miRNA binding to ceRNAs, leading to higher ceRNA expression, the co-expression analysis first computes the Pearson correlation coefficients between GOI expression levels and predicted ceRNAs by sequence-pairing and then removes the mRNAs with negative correlation coefficients. The p-values were calculated by Fisher transformation20 and the resultant predictions have two scores: those by sequence-pairing and those by co-expression test. We discuss their consolidation in the next section.
Score consolidation
To fuse these two scores, we utilized the Borda counting method21, which essentially sums ranks of scores. The resultant ceRNAs list can be downloaded from the website as a common delimited text file that contains the gene symbols ordered based on the Borda scores from highest to lowest, their sequence-pairing scores, co-expression scores, and their rankings.
Generation of regulatory network based on a GOI
TraceRNA also aims to provide a tool that allows biologists to discover new regulatory networks that are potentially modulated by a set of GTmiRs and gain insight into this novel gene regulation-modulation mechanism. To generate a GOI-ceRNA regulatory network, the user can select top predicted ceRNAs by co-expression test for a given GOI and then treat each predicted ceRNA as a new GOI (or cGOI). TraceRNA performs new rounds of predictions for each cGOI iteratively using the same number of predicted miRNAs that target each cGOI as described before. The resulting list (containing GOI, ceRNAs, and scores for all cGOIs) is used to generate a regulation network using Cytoscape plug-in22 and it is saved as a file that can be downloaded for further analysis.
Biological Functional Enrichment
To examine the functional association of ceRNAs for a given GOI, we used DAVID23 (http://david.abcc.ncifcrf.gov/), which uses a modified Fisher’s exact test to evaluate the functional enrichment of 40 annotation categories, including GO terms, protein-protein interactions, disease associations, pathways, homologies and other gene sets in a given gene list. In this paper, the enrichment results for p-value < 0.01 are reported.
Final remarks on methods
Discussion of TraceRNA implementation can be found in Supplemental Material. Table 1 summarizes the algorithms and databases used in TraceRNA. MiRTarBase, SVMicrO, and BCMicrO were implemented as databases queried by SQL commands. SiteTest accesses SVMicrO database and calculates binding scores for each ceRNA. All the computations including statistical significance and Borda fusion were implemented by R (http://www.r-project.org/).
Table 1.
Summary of TraceRNA Algorithms
| Algorithm or Database | Output | Function of Algorithm |
|---|---|---|
| miRTarBase | Experimentally verified miRNA:target interactions | Identify validated miRNAs |
| SVMicrO | Scores of miRNA-gene binding prediction by using a large number of binding site and 3’UTR features | Predict miRNAs binding |
| BCMicrO | Scores of miRNAs-genes binding predictions by fusion of 6 algorithms | |
| SiteTest | Scores of miRNAs-genes binding predictions using 3’UTR features | |
| ceRNA-SVMicrO ceRNA-BCMicrO ceRNA-SiteTest |
Scores for predicted ceRNAs by sequence-pairing | Predict ceRNAs by sequence-pairing |
| Expression Correlation | Pearson correlation of GOI and predicted ceRNAs by sequence-pairing | Calculate correlation between predicted ceRNAs by sequence-pairing |
| SeqExp Fusion (Borda merging method) | Scores from fusing sequence-level predictions and expression correlation | Fuse sequence-level predictions and co-expression |
| ceRNA-Net | List of gene pairs (nodes) and scores of their directed interactions | Discover regulatory networks |
Results and Case Studies
TraceRNA integrates databases, sql queries, real-time predictions, and generation of ceRNA interaction network under a unified web interface, enabling ceRNA predictions and discovery of novel biological regulation. We illustrate its features and capabilities next.
TraceRNA web interface
The TraceRNA web interface (Fig. S1), starts with a query GOI by the user. Currently, TraceRNA only supports official gene symbols from the UCSC annotation. Given a GOI, a set of validated miRNAs that target the GOI derived from miRTarBase will be displayed under the checker box "Select validated miRNAs." Other miRNAs predicted by SVMicrO are listed under "Select Predicted miRNAs" in an increasing order of binding p-values of targeting GOI. The user can select from these two sources a set of miRNAs to form GTmiRs. A rule of thumb: one can require binding p-value<0.01, which is expected to produce less than 43 miRNAs for 50% of genes (Fig. S2), or miRNAs log2 expression level in GBM > 6 (Fig. S3).
After selection, the user can choose from SVMicrO, BCMicrO, or SiteTest and further integrate gene expression data. To evaluate ceRNA-mediated gene-gene interactions, the user can perform ceRNA prediction iteratively by treating top K (20 by default) ceRNAs as GOI, (cGOI). The resulting interactions will be displayed within the web-interface and can be also saved in a file to be imported into Cytoscape.
GTmiR determination
Determination of GTmiRs that target the GOI is an important step that can significantly affect the final prediction. GTmiRs are ideally determined by experiments, but the complete set of experimentally validated GTmiRs are rarely available. ceRNA predictions based on a subset of validated GTmiRs will have low specificity and predicted GTmiRs are needed to increase the specificity. However, high false positives associated with current miRNA target prediction algorithms could introduce false positives in ceRNA predictions, thus potentially harming rather than improving the ceRNA predictions specificity. Furthermore, a different number of candidate GTmiRs can also affect the prediction performance, where a lower number will likely produce lower prediction specificity, whereas a number too high can, on the contrary, harm the prediction sensitivity. This observation was captured in Supplemental Fig. S4, in which we varied the number of miRNAs from 2 to 70. It clearly demonstrated the low specificity with a small number of miRNAs and a loss of sensitivity when too many miRNAs were selected. Therefore, care needs to be taken in choosing GTmiRs. To this end, TraceRNA provides flexibility to choose between validated and predicted GTmiRs or a combination of both.
As an example, Table 2 includes the top 20 experimentally validated (from miRTarBase) and predicted GTmiRs for PTEN, ESR1, and BRCA1, respectively. In all three cases, the numbers of experimentally validated GTmiRs are less than 20, and there are only 4 for BRCA1. Apparently, using the validated GTmiRs alone will result in low specificity in ceRNA predictions.
Table 2.
Top 20 experimentally validated and predicted GTmiRs for PTEN, ESR1 and BRCA1.
| PTEN | ESR1 | BRCA1 | |||
|---|---|---|---|---|---|
| 20 predicted by SVMicrO |
miRTarBase | 20 predicted by SVMicrO |
miRTarBase | 20 predicted by SVMicrO |
miRTarBase |
| miR-16 | miR-106b | miR-518c | miR-18a | miR-1915 | miR-15a |
| miR-512-3p | miR-141 | miR-558 | miR-18b | miR-588 | miR-16 |
| miR-30c-1* | miR-17 | miR-1914 | miR-193b | miR-324-3p | miR-212 |
| miR-142-5p | miR-18a | miR-1305 | miR-19a | miR-29b-2* | miR-24 |
| miR-323-3p | miR-19a | miR-551b* | miR-19b | miR-548p | |
| miR-548c-3p | miR-19b | miR-934 | miR-206 | miR-1304 | |
| miR-143 | miR-20a | miR-509-5p | miR-20b | miR-1913 | |
| miR-580 | miR-21 | miR-142-3p | miR-22 | miR-345 | |
| miR-26a | miR-214 | miR-1233 | miR-221 | miR-545 | |
| miR-510 | miR-216a | miR-1915 | miR-222 | miR-498 | |
| miR-212 | miR-217 | miR-582-3p | miR-29b | miR-146a | |
| miR-26b | miR-221 | miR-590-3p | miR-302c | miR-30e* | |
| miR-194 | miR-222 | miR-302a* | let_7g* | ||
| miR-202* | miR-26a | miR-605 | miR-1914 | ||
| miR-494 | miR-494 | miR-647 | miR-1910 | ||
| miR-1253 | miR-1913 | miR-760 | |||
| miR-340 | miR-34a* | miR-615-5p | |||
| miR-410 | miR-362-3p | miR-9 | |||
| miR-335* | miR-629 | miR-516a-3p | |||
| miR-513a-3p | miR-34b | miR-516b* | |||
Significance of ceRNA Prediction Score
Fig. 2-(a) depicts the empirical distribution of the genome-wide SVMicrO scores for 772 human miRNAs and a mixture of two distributions can be clearly observed, one clustered around smaller scores with a much larger mass and the other around larger scores. As discussed in the Methods section, the peak around smaller scores was considered to represent the null distribution and was fitted with a Gamma distribution, whose parameters are α=0.7234 and β=0.3594 with 95% confidence interval (0.7229, 0.7239) and (0.3591, 0.3598) for α and β, respectively. Fig. 2-(b) shows the histogram and the fitted Gamma distribution (with a constant shift). Table S1 lists the fitted parameters of the Gamma distributions for SVMicrO, BCMicrO and SiteTest.
Case Study 1
We applied TraceRNA to predict the ceRNAs of PTEN. PTEN is a gene related to the development of many cancers, where it often functions as a key tumor suppressor, whose abundance determines the critical outcomes in tumorigenesis6. PTEN is also known to regulate cell cycle, particularly in preventing cells from growing and dividing too rapidly. PTEN ceRNAs have also been predicted and reported6, 7.
To predict ceRNAs of PTEN with TraceRNA, we selected only the predicted miRNAs as GTmiRs (Table 2) and further chose SVMicrO to predict the ceRNAs by sequence-pairing. TraceRNA returned a total of 761 predicted ceRNAs by sequence-pairing for p-value < 0.05, and the 20 best predictions (in descending order of prediction score) together with p-values are provided in Table 3a. One important feature of TraceRNA is the possibility to increase the predictions specificity and predict context-specific ceRNAs by integrating an expression dataset of a disease condition. In this case, 400 GBM expression samples from TCGA project were included, based on which co-expression correlations against PTEN were evaluated and a total of 466 genes were obtained for Pearson correlation greater than zero as the GBM-specific ceRNAs of PTEN. Table 3b shows the results for the top 20 predicted ceRNAs based on Borda score. To examine their functions, pathway enrichment was performed using DAVID23 on these 466 ceRNAs, and the 3 enriched pathways are shown in Table 4 (under TraceRNA+GBM). Well known cancer related pathways including MAPK signaling and WNT signaling were significantly enriched, indicating an important involvement of PTEN ceRNAs in cancer. Another enriched pathway, TGF- signaling, is known to utilize intracellular SMADs to mediate growth suppression and PTEN down-regulation simultaneously to induce growth proliferation. Here, the prediction result provided a third possible regulatory mechanism of TGF regulation by PTEN via its ceRNAs.
Table 3.
Predicted ceRNAs of PTEN
| A: TraceRNA (sequence-based) | B:TraceRNA (w/ GBM Gene Expression) | C: ceRDB | ||||||
|---|---|---|---|---|---|---|---|---|
| Symbol | SVMicrO score |
p-value | Symbol | SVMicrO score |
p-value | Correlation coefficient |
Symbol | Score |
| QKI | 1.199 | 0.0031 | GSPT1 | 0.9105 | 0.0065 | 0.5534 | TNRC6B | 26 |
| CPEB2 | 1.099 | 0.0040 | CD2AP | 0.8590 | 0.0074 | 0.5632 | TET3 | 25 |
| OTUD4 | 1.079 | 0.0042 | USP15 | 1.0550 | 0.0045 | 0.5167 | NFIB | 23 |
| PPP6C | 1.071 | 0.0043 | PPP6C | 1.0706 | 0.0043 | 0.4913 | ATXN1 | 21 |
| NOVA1 | 1.059 | 0.0044 | RAD23B | 0.9124 | 0.0065 | 0.5167 | ANKRD52 | 20 |
| USP15 | 1.055 | 0.0045 | NIPBL | 0.7567 | 0.0097 | 0.6163 | tcag7.1228 | 20 |
| LEMD3 | 1.031 | 0.0048 | CNOT6 | 0.8730 | 0.0072 | 0.4888 | NUFIP2 | 17 |
| SUZ12 | 1.027 | 0.0048 | ARF6 | 0.7706 | 0.0093 | 0.5561 | CPEB4 | 16 |
| NARG1 | 1.022 | 0.0049 | MED14 | 0.8159 | 0.0083 | 0.5100 | ZFHX4 | 16 |
| ZFHX3 | 1.014 | 0.0050 | KIAA0240 | 0.9639 | 0.0057 | 0.4572 | BACH2 | 15 |
| C18orf25 | 1.009 | 0.0050 | SOCS5 | 0.7573 | 0.0096 | 0.5438 | CLCN5 | 15 |
| FNDC3B | 1.007 | 0.0051 | CSNK1G3 | 0.7361 | 0.0102 | 0.5675 | CUGBP2 | 15 |
| BCL11A | 0.967 | 0.0056 | NAB1 | 0.7867 | 0.0089 | 0.4986 | ONECUT2 | 14 |
| ST18 | 0.967 | 0.0056 | CBFB | 0.7516 | 0.0098 | 0.5273 | AFF1 | 14 |
| KIAA0240 | 0.964 | 0.0057 | RPS6KB1 | 0.7437 | 0.0100 | 0.5387 | BNC2 | 14 |
| RSBN1 | 0.963 | 0.0057 | RPS6KA3 | 0.7126 | 0.0108 | 0.5780 | KLF12 | 14 |
| TET2 | 0.961 | 0.0057 | TBL1XR1 | 0.7553 | 0.0097 | 0.5019 | NFAT5 | 14 |
| ICK | 0.952 | 0.0058 | RBM12 | 0.8482 | 0.0076 | 0.4516 | OTUD4 | 14 |
| SCN2A | 0.951 | 0.0058 | ELL2 | 0.8582 | 0.0074 | 0.4472 | ZBTB34 | 14 |
| PAN3 | 0.925 | 0.0062 | ZFX | 0.7595 | 0.0096 | 0.4752 | ZEB2 | 14 |
(a) Best 20 predicted ceRNAs by sequence-pairingof PTEN predicted by SVMicrO based on miRNAs in Table 2 (first column, top 20 predicted miRNAs).
(b) Top 20 predicted ceRNAs by co-expression testof PTEN under the context of GBM. Column “p-value” shows the prediction p-values obtained in (a). Column “Correlation coefficient” lists Pearson correlations between a ceRNA and PTEN calculated using GBM gene expression data (see Materials and Methods). The ranking is based on the Borda merging of the rankings in (a) and that based on the Pearson correlation. ceRNAs shared between Tables 3a and 3b are in b font.
(c)The ceRNAs obtained from ceRDB. ceRNAs that were predicted by both algorithms (within top 50) are in italic. Note: tcag7.1228 is a synonym of UBN2.
Table 4.
Enriched pathways of the predicted PTENceRNAs
| KEGG Pathways | TraceRNA | TraceRNA + GBM |
ceRDB |
|---|---|---|---|
| Long-term potentiation | 0.00002 | 8.8×10−8 | 0.00844 |
| Oocyte meiosis | 0.00066 | 1.9×10−6 | |
| MAPK signaling pathway | 0.00239 | 0.00096 | |
| Wnt signaling pathway | 0.00496 | 0.00005 | |
| Neurotrophin signaling pathway | 0.00019 | ||
| Axon guidance | 0.00460 | 0.00998 | |
| Insulin signaling pathway | 0.00073 | ||
| TGF-beta signaling pathway | 0.00036 | 0.00543 | |
| ErbB signaling pathway | 0.00110 | ||
| Endocytosis | 0.00210 | ||
| Ubiquitin mediated proteolysis | 0.00180 | ||
| Focal adhesion | 0.00074 | ||
| Melanogenesis | 0.00390 |
The table includes the top enriched (p-values <0.01) pathways for three ceRNA prediction results. Enrichment analysis was performed by DAVID with entire human genome as the background gene set.
It would be also interesting to examine if these GBM specific ceRNAs by co-expression test also have higher specificity than those by sequence-pairing alone (Table 3a). However, direct comparison was infeasible due to a lack of true PTEN ceRNAs. Alternatively, pathway enrichment of the predictions result was conducted to make an indirect comparison. Intuitively, true ceRNAs should be functionally more significant than false positive predictions, and therefore, the predictions with higher specificity should be accompanied by a larger number of more enriched pathways. Pathway enrichment of sequence-based predictions is shown in Table 4 (under TraceRNA) and it is apparent that the GBM-specific ceRNAs by co-expression test are of higher functional enrichment (9 enriched functions vs 4 weakly enriched functions in sequence-pairing predictions alone), thus a higher predictions specificity. Expression (GBM data) scatter-plots of PTEN vs. three predicted ceRNAs (QKI, NOVA1, and BCL11A) by sequence predictions (Table 3a) are shown in Fig. 3(a)–(c). The correlations are clearly very low, suggesting that they are not GBM-specific ceRNAs. As expected, they were not among the predicted GBM-specific ceRNAs (Table 3b). Expression scatter-plots of PTEN vs. the 3 predicted GBM-specific ceRNAs (GSPT1, PPP6C, and USP15; Table 3b) are shown in Fig. 3(d)–(f). Their correlations are much higher. Notice that USP15 was also ranked No. 6 in sequence-based predictions. As expected, its ranking improved after integrating gene expression data.
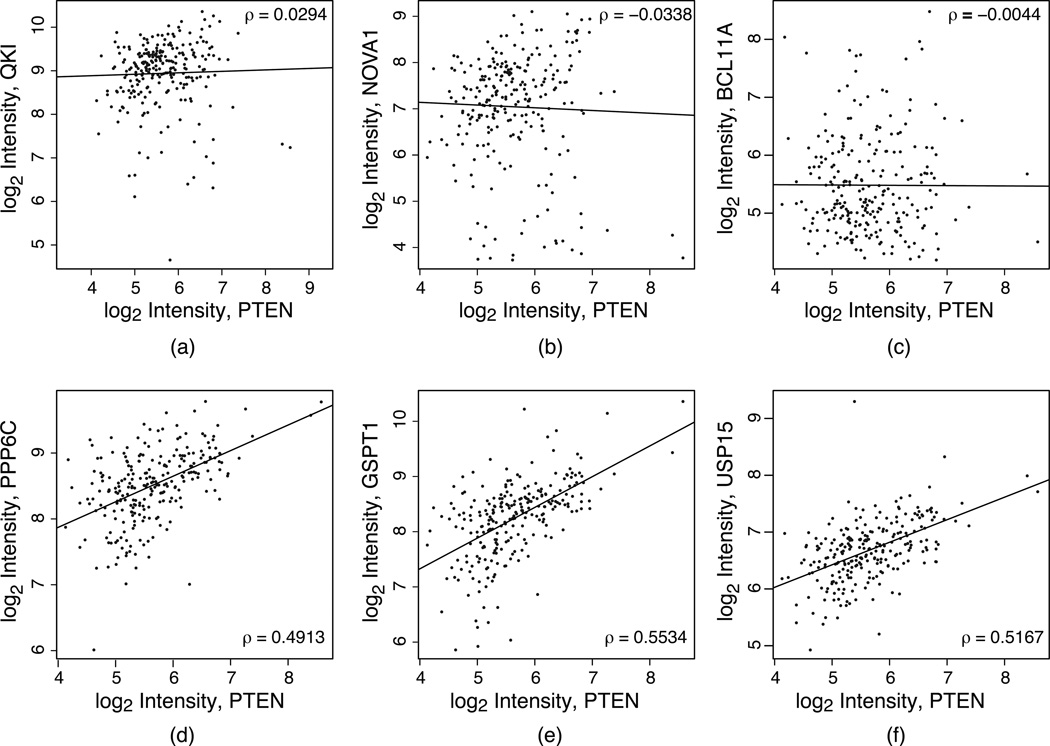
Figure 3.
Expression scatter plots of PTEN vs. top predicted ceRNAs in GBM. (a)–(c) The three genes were top ranked ceRNAs predicted by sequence-pairing listed in Table 3a. However, their gene expression correlations to GOI (PTEN) are low and insignificant. Consequently, they were not among the top ranked ceRNAs after considering co-expression with PTEN (see Table 3b). (d)–(e) Three top ranked GBM-specific ceRNAs predicted by sequence-pairing and co-expression test (Table 3b). Clearly, their higher correlation coefficients enhance their prediction p-values.
As a comparison, ceRNAs predicted by Competitive Endogenous mRNA DataBase (ceRDB)24 were also retrieved and top 20 are listed in Table 3c. We observed only 1 overlaps between ceRDB and TraceRNA predictions in the top 20 predictions. To examine the functional significance of the ceRDB predictions, pathway enrichment was conducted (Table 4, column ceRDB). 9 pathways in TraceRNA+GBM are enriched compared to 5 pathways in ceRDB. For example, enrichment p-values of “Long-term Potentiation” that includes genes such as MAPK1, NRAS, RPS6KA3, KRAS, and CREBBP, are 8.8×10−8, 0.00002 and 0.0084 for TraceRNA+GBM, TraceRNA sequence-pairing alone, and ceRDB, respectively.
Case Study 2
Breast cancer (BC) is a common disease in women and its incidence is still increasing25 despite great improvement in therapies and earlier screening26. Hormone receptor (such as estrogen receptor, ER) ERpositive breast cancer and ERBB2-positive breast cancer (about 50% co-expressed with ER+ tumors) currently account for about 75% and 15% of all breast cancer cases, respectively. The remaining 10% are so-called triple-negative breast cancers (TNBC), as defined by absent expression of ER, progesterone receptor (PR) and ERBB2 proteins27, 28. In this study, our objective is to identify genes mediated by the estrogen receptor alpha, ESR1, through ceRNA regulatory network in breast cancer. To this end, we selected ESR1 as the GOI, and then predicted miRNAs as GTmiRs (Table 2, column 2). A total of 730 predicted ceRNAs by sequence-pairing were obtained by SVMicrO at p-value<0.05. Top 20 predictions were provided in Table 5a. Predicted BC-specific ESR1 ceRNAs by co-expression test were subsequently obtained by including TCGA gene expression of 590 breast cancer tumor samples (described in Materials and Methods). A total of 378 BC-specific ceRNAs were obtained and top 20 are shown in Table 5b.
Table 5.
Predicted ceRNAs of ESR1
| A. TraceRNA (sequence-based) | B. TraceRNA (w/ BRCA Gene Expression) | |||||
|---|---|---|---|---|---|---|
| Symbol | SVMicrO Score |
p-value | Symbol | SVMicrO score |
p-value | Correlation |
| RUNX1 | 0.9158 | 0.0064 | FOXP1 | 0.6874 | 0.0116 | 0.5082 |
| MAP2 | 0.8791 | 0.0070 | NRIP1 | 0.6654 | 0.0122 | 0.5419 |
| MIER3 | 0.8748 | 0.0071 | MIER3 | 0.8748 | 0.0071 | 0.3434 |
| CTDSPL2 | 0.8649 | 0.0073 | CPEB3 | 0.6562 | 0.0125 | 0.4838 |
| LOC440354 | 0.7936 | 0.0088 | ZBTB4 | 0.6587 | 0.0124 | 0.4226 |
| LOC595101 | 0.7923 | 0.0088 | ARL3 | 0.5801 | 0.0153 | 0.5627 |
| DDX3Y | 0.7771 | 0.0092 | RUNX1 | 0.9158 | 0.0064 | 0.2674 |
| GUCY1B2 | 0.7674 | 0.0094 | FCHO2 | 0.5846 | 0.0151 | 0.3999 |
| RNF44 | 0.7628 | 0.0095 | NOVA1 | 0.5455 | 0.0167 | 0.4788 |
| CTTNBP2NL | 0.7586 | 0.0096 | BSN | 0.5917 | 0.0148 | 0.3294 |
| C20orf112 | 0.7572 | 0.0097 | OTUD7B | 0.6230 | 0.0137 | 0.2962 |
| WAPAL | 0.7551 | 0.0097 | MAX | 0.5842 | 0.0151 | 0.3260 |
| SKI | 0.7352 | 0.0102 | ALCAM | 0.5379 | 0.0170 | 0.4151 |
| E2F3 | 0.7311 | 0.0103 | CTDSPL | 0.5544 | 0.0163 | 0.3245 |
| NIPBL | 0.7307 | 0.0103 | SP1 | 0.6709 | 0.0121 | 0.1925 |
| UBE2Z | 0.7168 | 0.0107 | KCNMA1 | 0.5680 | 0.0157 | 0.2949 |
| G3BP2 | 0.7061 | 0.0110 | STRBP | 0.5903 | 0.0149 | 0.2605 |
| MYT1L | 0.7050 | 0.0110 | BTRC | 0.4881 | 0.0194 | 0.5566 |
| DDX3X | 0.7029 | 0.0111 | MYST3 | 0.6165 | 0.0139 | 0.1916 |
| KIAA0240 | 0.6981 | 0.0112 | POMGNT1 | 0.5768 | 0.0154 | 0.2208 |
(a) Best 20 predicted ceRNAs by sequence-pairing of ESR1 predicted by SVMicrO alone using top 20 miRNAs in Table 3b. ceRNAs are sorted by SVMscore.
(b) BRCA-specific ceRNAs obtained by integrating predictions in Table 5a) with TCGA breast cancer gene expression. Rows in italic are cross-listed in both (a) and (b).
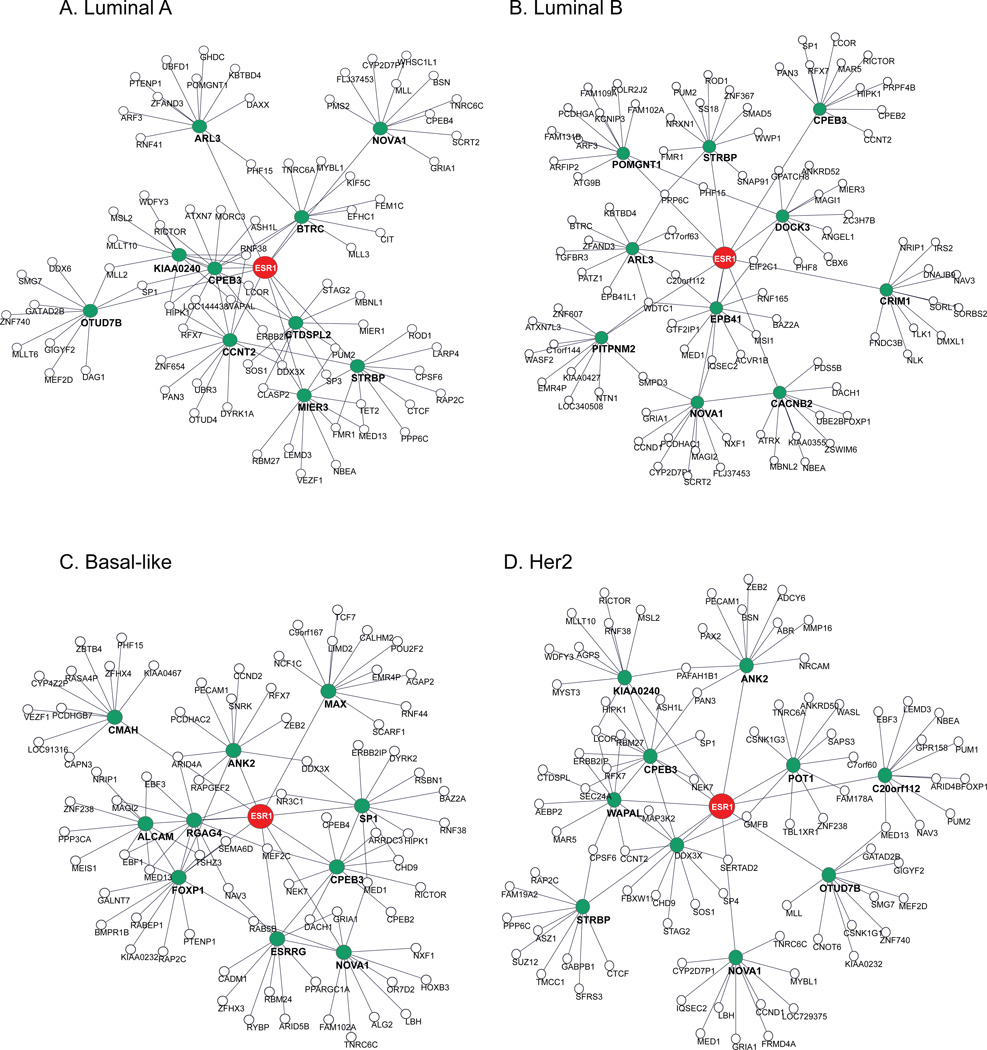
To substantiate our finding, we examined the gene regulation networks modulated by ESR1 ceRNAs in different breast cancer subtypes. As classified by earlier studies19, 4 major subtypes, determined by molecular signatures, are luminal A (ER+, PR+, Her2−), luminal B (ER+, PR+, Her2+), basal-like (mostly TNBC), and Her2 (amplified or over-expressed ERBB2). To construct subtype-specific ESR1 mediated ceRNA networks, we prepared 4 TCGA expression datasets for the corresponding 4 subtypes, which included 93, 56, 228, and 123 samples for Basal-like, HER2, Luminal A and Luminal B, respectively. Considering that genes may express constantly within each subtype, we added all normal reference samples to each subtype to increase the dynamic range for correlation analysis. For each subtype, co-expression analysis was performed and integrated with sequence-paring predictions (Table 5a). To generate the interaction network, the process was repeated on top 10 predicted ceRNAs. Figs. 4A–4D illustrate the resulting subtype-specific ceRNA networks. Among these networks, only 2 first layer ceRNAs (NOVA1 and CPEB3) are shared. NOVA1, neuro-oncological ventral antigen 1, has been implicated in breast cancer29 and shown correlated in gene expression with ESR130, and CPEB3, a regulator of EGFR31, has been shown to be important in breast cancer32. While these two genes’ expression levels are mediated by ESR1 via GTmiRs in all four subtypes, other unique ceRNAs are also very important to each subtype. For example, ceRNA MAX in Basal-like regulation (Fig. 4C) is an important partner of proto-oncogene Myc in driving cell proliferation in variety of tumors. In the case of Her2 (Fig. 4D), ESR1 interacts with ANK2 and then PAX2, another gene that plays critical role in breast cancer33.
Figure 4.
ESR1 ceRNA interactions networks for four breast cancer subtypes. Each network consists of three layers. The top layer includes the GOI (the largest node), the second layer includes 10 ceRNAs (medium size nodes) predicted in the first iteration, and the third layer includes predictions (smallest nodes) from the second iterations.
Discussion
Here we presented TraceRNA, an easy to use web application for predictions of ceRNAs of a GOI and their interaction network. This web application is motivated by lack of ready-to-use tools for ceRNA predictions and, to the best of our knowledge, TraceRNA is the only web application other than ceRDB that is specialized for ceRNA predictions. Compared with ceRDB, TraceRNA has richer functionality designed to meet different research needs. Because of the high false positive rate and low sensitivity of existing miRNA target prediction algorithms, TraceRNA provides the users with 3 different algorithms so that they can compare and/or complement results as a remedy to the potentially poor predictions from a single algorithm. TraceRNA also includes the validated targets from mirTarBase, which can be selected to potentially improve the specificity of ceRNA predictions. TraceRNA also enables predictions of context-specific ceRNAs by integrating co-expression with sequence-level predictions. In the current version, two expression datasets from TCGA have been preloaded into the web database. All prediction results include p-values and FDR as statistical significance. What is also unique about TraceRNA is its ability to construct and plot ceRNA interaction networks. This network can reveal important interactions that might not be easily perceived with the list of predicted ceRNAs. The generated network plots can be downloaded and are ready to be used for scientific publication. In contrast, ceRDB includes only one algorithm for miRNA target predictions and is also devoid of the afore-mentioned functions in TraceRNA.
Two case studies, prediction of PTEN ceRNAs and that of ESR1 ceRNAs, were presented to demonstrate the effectiveness of TraceRNA in making biological meaningful predictions. Because both genes are important cancer associated genes, their ceRNAs in the context of cancer were also predicted. In the case of PTEN, the GBM specific ceRNAs were shown to be functionally more enriched than the sequence-level predictions alone and important signaling pathways known to be related with PTEN regulation were also predicted among the most enriched pathways. When compared with ceRDB, TraceRNA predictions were functional much more enriched, indicative of higher predictions specificity. For ESR1, unique ceRNA interaction networks for four breast cancer subtypes were constructed. While ceRNAs common to four networks were observed, considerable differences exist among these four networks in ceRNAs and their interactions. Examples were provided to show possible links between the unique ceRNA interactions and the subtypes, which suggests that these differences in ceRNA interactions may very well be used to explain the genomics mechanisms underlying the subtypes. If proven true, the ceRNA networks could provide an alternative to the genomics markers for disease treatment. Taken together, TraceRNA has been shown as an effective tool for context-specific ceRNA predictions and discovery of ceRNA interactions modulated by GTmiRs.
Context-specific ceRNAs are closely dependent on miRNA expressions. A predicted ceRNA by sequence-pairing could not compete with GOI for binding of weakly expressed miRNAs. As a result, only highly expressed miRNAs should be considered in ceRNA predictions. Current version of TraceRNA does not yet consider miRNA expression for ceRNA predictions but displays expression values to help the user to select GTmiRs. On the other hand, mRNA expression profiles are still much more accessible than miRNA expression data, ceRNA predictions based on mRNA expression will still be of higher interest in practice. However, as miRNA profiles become increasingly available, there will be more demand to include miRNA expression in ceRNA predictions to achieve more accurate context-specific predictions. Future work should allow us to incorporate this function in TraceRNA to enable predictions in a miRNA expression dependent fashion.
Supplementary Material
Acknowledgments
TraceRNA is freely accessible at http://compgenomics.utsa.edu/cerna. As a web application, there is no requirement for the users to access the application other than internet connection and a browser.
Funding Sources: This work is supported in part by a National Science Foundation grant (CCF-0546345), a Qatar National Research Fund grant (09-874-3-235), and National Institute of Health grants (NIH-NCATS UL1TR000149 and U54 CA11300126, Integrative Cancer Biology Program)..
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, et al. A synonymous variant in irgm alters a binding site for mir-196 and causes deregulation of irgm-dependent xenophagy in crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 3.Kozomara A, Griffiths-Jones S. Mirbase: Integrating microrna annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medina PP, Slack FJ. Micrornas and cancer: An overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 5.Yue D, Meng J, Lu M, Chen P, Guo M, Huang Y. Understanding microrna regulation: A computational perspective. IEEE Signal Process Magazine. 2012;29:77–88. [Google Scholar]
- 6.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A cerna hypothesis: The rosetta stone of a hidden rna language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Califano A, et al. An extensive microrna-mediated network of rna-rna interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu S-D, Lin F-M, Wu W-Y, Liang C, Huang W-C, Chan W-L, et al. Mirtarbase: A database curates experimentally validated microrna-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Yue D, Chen Y, Gao SJ, Huang Y. Improving performance of mammalian microrna target prediction. BMC Bioinformatics. 2010;11:476. doi: 10.1186/1471-2105-11-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue D, Guo M, Chen Y, Huang Y. A bayesian decision fusion approach for microrna target prediction. BMC Genomics. 2012;13(Suppl 8):S13. doi: 10.1186/1471-2164-13-S8-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Microrna targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microrna.Org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microrna target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 14.Wang X. Mirdb: A microrna target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microrna target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 16.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, et al. Diana-microt web server: Elucidating microrna functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y BYaH. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57 [Google Scholar]
- 18.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotelling H. New light on the correlation coefficient and its transforms. J Roy Stat Soc B. 1953;15:193–232. [Google Scholar]
- 21.Montague JAAaM. Models for metasearch. ACM SIGIR Special Interest Group on Information Retrieval 2001 New Orleans, LA, 2001. 2001 [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The david gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarver AL, Subramanian S. Competing endogenous rna database. Bioinformation. 2012;8:731–733. doi: 10.6026/97320630008731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 26.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, et al. Cancer I, Surveillance Modeling Network C. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the carolina breast cancer study. Clin Cancer Res. 2010;16:6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 29.Buckanovich RJ, Darnell RB. The neuronal rna binding protein nova-1 recognizes specific rna targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Peng SC, Lai YT, Huang HY, Huang HD, Huang YS. A novel role of cpeb3 in regulating egfr gene transcription via association with stat5b in neurons. Nucleic Acids Res. 2010;38:7446–7457. doi: 10.1093/nar/gkq634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitteri SJ, Amon LM, Busald Buson T, Zhang Y, Johnson MM, Chin A, et al. Detection of elevated plasma levels of epidermal growth factor receptor before breast cancer diagnosis among hormone therapy users. Cancer Res. 2010;70:8598–8606. doi: 10.1158/0008-5472.CAN-10-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harari D, Yarden Y. Molecular mechanisms underlying erbb2/her2 action in breast cancer. Oncogene. 2000;19:6102–6114. doi: 10.1038/sj.onc.1203973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.