Abstract
In vivo aggregation of tau protein is a hallmark of many neurodegenerative disorders, including Alzheimer’s disease (AD). Recent evidence has also demonstrated activation of the unfolded protein response (UPR), a cellular response to endoplasmic reticulum (ER) stress, in AD, although the role of the UPR in disease pathoge-nesis is not known. Here, three model systems were used to determine whether a direct mechanistic link could be demonstrated between tau aggregation and the UPR. The first model system used was SH-SY5Y cells, a neuronal cultured cell line that endogenously expresses tau. In this system, the UPR was activated using chemical stressors, tunicamycin and thapsigargin, but no changes in tau expression levels, solubility, or phosphorylation were observed. In the second model system, wild-type 4R tau and P301L tau, a variant with increased aggregation propensity, were heterologously overexpressed in HEK 293 cells. This overexpression did not activate the UPR. The last model system examined here was the PS19 transgenic mouse model. Although PS19 mice, which express the P301S variant of tau, display severe neurodegeneration and formation of tau aggregates, brain tissue samples did not show any activation of the UPR. Taken together, the results from these three model systems suggest that a direct mechanistic link does not exist between tau aggregation and the UPR.
Keywords: unfolded protein response, Alzheimer’s disease, SH-SY5Y, PS19, neurofibrillary tangles, paired helical filaments
Alzheimer’s disease (AD) is the most prevalent form of dementia; approximately 5.3 million Americans currently have AD, and the number increases each year as the average age in our society increases (Alzheimer’s Association, 2009). Pathologically, the disease involves neuronal death accompanied by accumulation of two distinct types of protein aggregates, extracellular aggregates of amyloid β (Aβ) peptide, often referred to as plaques, and intracellular aggregates of tau protein, often referred to as neurofibrillary tangles (NFTs) or paired helical filaments (PHFs; Selkoe, 2001). It is unclear how Aβ aggregates and tau aggregates are related and which is more important in disease pathology (Lee, 2001).
Tau protein is a natively unfolded protein, abundant in neurons, which binds to and stabilizes microtubules. Tau has 79 potential phosphorylation sites, and its phosphorylation is regulated by phosphatases and kinases, including glycogen synthase kinase-3β (GSK-3β). Hyperphosphorylated tau is associated with the disease state, insofar as it shows decreased microtubule binding and an increased propensity for tangle formation (Buee et al., 2000). In contrast to Aβ aggregation, which is specifically associated with AD, tau aggregates form in multiple neurodegenerative disorders, collectively termed tauopathies, including Pick’s disease, dementia pugilistica, and frontaltemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17; Hutton et al., 1998; Lee, 2001; Brandt et al., 2005).
An increasing amount of evidence indicates that endoplasmic reticulum (ER) stress may play a significant role in AD. The ER is the first organelle in the secretory pathway of eukaryotic cells and is responsible for ensuring proper folding of newly synthesized proteins. If there is an accumulation of unfolded proteins in the ER, a set of signaling pathways known as the unfolded protein response (UPR) is activated. This adaptive response attempts to restore ER homeostasis by decreasing overall protein production while selectively increasing production of chaperones and proteases. If ER stress persists, the UPR will eventually induce apoptosis or programmed cell death (Cudna and Dickson, 2003; Szegezdi et al., 2003; Schroder and Kaufman, 2005; Lai et al., 2007; Malhotra and Kaufman, 2007). The mammalian UPR consists of three signaling pathways, each induced by an ER-membrane-resident stress sensor, inositol-requiring kinase 1 (IRE1), protein kinase-like ER kinase (PERK), or activating transcription factor 6 (ATF6; Wu and Kaufman, 2006; Malhotra and Kaufman, 2007).
Given the important role of the ER in protein synthesis and quality control as well as the potential for ER stress to lead to apoptosis, it is not surprising that ER dysfunction may play a role in AD. Neuronal tissue from AD patients shows multiple UPR activation markers, including elevated levels of BiP (Hoozemans et al., 2005), increased phosphorylation of PERK (Hoozemans et al., 2005, 2009), increased phosphorylation of eIF2α (Hoozemans et al., 2009), and disruption of ER Ca2+ regulation (Verkhratsky, 2005), indicating that the UPR is activated in AD.
ER stress could potentially provide a link between extracellular Aβ aggregation and intracellular tau aggregation. Treatment of cultured neuronal cells with Aβ aggregates has been shown to cause both UPR induction (Chafekar et al., 2007; Resende et al., 2008) and increased phosphorylation of tau (Resende et al., 2008), although the mechanism by which Aβ causes these effects is unclear. GSK-3β, a known tau kinase, is activated by the UPR; thus, ER stress could be a cause of increased tau phosphorylation. Chronic ER stress resulting in ER-induced apoptosis has been suggested as a potential cause of neuronal death in AD (Katayama et al., 2004), but it is also possible that the UPR plays a neuroprotective role.
Although AD patients clearly exhibit ER stress, it remains unclear what role this stress plays in the disease. Is ER stress caused by formation of Aβ aggregates or tau aggregates or both? Does activation of the UPR result in increased aggregate formation? Is the UPR a cause of neuronal cell death? The current work uses well-controlled model systems in an attempt to understand better the potential mechanistic connections between the UPR and tau dysfunction.
MATERIALS AND METHODS
Materials
All chemicals used were obtained from major chemical suppliers. Unless otherwise noted, all cell culture reagents were obtained from Invitrogen (Carlsbad, CA).
Cell Culture
SH-SY5Y cells were obtained from the American Type Culture Collection (ATCC). Cells were grown in a 1:1 mixture of Eagles’s minimum essential medium (EMEM) and Ham’s F12 medium supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 µg/ml streptomycin at 37°C and 5% CO2. Medium was replenished every 2–3 days. Cells were maintained in T-75 flasks and passaged at a ratio of 1:20 to 1:50 when flasks reached 80–90% confluence.
SH-SY5Y cells were differentiated with retinoic acid for all experiments. Cells were removed from T-75 flasks with 0.25% trypsin and 0.5 mM EDTA, and cell concentration was determined by hemocytometry. Cells were plated in six-well plates at a density of 5 × 104 cells/well. Approximately 5–7 days after plating, medium was supplemented with 10 µM retinoic acid. Cells were cultured in the presence of retinoic acid for a minimum of 8 days prior to use in UPR experiments.
Human embryonic kidney cells (HEK293) stably trans-fected with a plasmid containing the tetracycline (Tet) repressor were obtained from Invitrogen and are referred to by the commercial name “T-REx-293.” T-REx-293 cells were cultured in Dulbecco’s modification of Eagles’s minimum essential medium (DMEM) with 4 mM L-glutamine and high glucose supplemented with 10% FBS, 50 U/ml penicillin, and 50 µg/ml streptomycin at 37°C and 5% CO2. Medium was replenished every 2–3 days. Media also included 5 µg/ml blasticidin, the selective marker for the Tet repressor plasmid. Cells were maintained in T-75 flasks and passaged at a ratio of 1:5 to 1:8 when flasks reached 80–90% confluence.
Transfection
T-REx-293 cells were transfected with 0.5 µg/well (in six-well plates) pT-REx-DEST31 Gateway vector (Invitrogen) containing the gene for wild-type tau (4R isoform), tau P301L, or a control gene (lacZ) using the reagent Lipofectamine 2000 (Invitrogen). Some cells were also treated with Lipofectamine in the absence of DNA as a negative control for transfection.
Cells were allowed to grow in antibiotic-free media for a 48-hr recovery period after transfection before switching to selective media including 5 µg/ml blasticidin and geneticin in varying concentrations (0, 200, 500, and 800 µg/ml). After approximately 7 days of selection, nontransfected control cells grown with 500 and 800 µg/ml geneticin concentrations were no longer viable. Stably transfected cells were then propagated in 500 µg/ml geneticin.
Chemical Induction of the UPR
To determine the effect of the UPR on tau biology, the UPR was induced using chemical reagents in differentiated SH-SY5Y cells, which natively express tau. To induce the UPR, medium containing 10 µg/ml tunicamycin or 500 nM thapsigargin was added to differentiated cells in six-well plates. As a positive control for tau phosphorylation, some cells were also treated with 75 nM okadaic acid, a phos-phatase inhibitor (Perez et al., 2002; Xu et al., 2004). At specified times after treatment, cell samples were taken and analyzed for both UPR activity and changes in tau phospho-rylation state and solubility.
Recombinant Protein Expression
T-REx-293 cells stably transfected with the DEST31 plasmid containing the gene of interest as well as nontransfected control cells were plated in six-well plates at a density of 2 × 105 cells/well. Cells were grown in selective media for ~24 hr before protein expression was induced by addition of 1 µg/ml tetracycline. At specified times after treatment, cell samples were taken for analysis by RT-PCR and Western blotting.
Mouse Tissue Samples
Brain samples from wild-type and PS 19 transgenic mice, which express P301S tau (Yoshiyama et al., 2007), collected at 6 and 11 months were a generous gift from Dr. Virginia Lee of the University of Pennsylvania’s Center for Neurodegenerative Disease Research. PS 19 mice have a neurodegenerative phenotype with limb weakness and brain atrophy after 3 months of age, progressing to paralysis at 7–10 months of age. In brain tissue samples from PS19 mice, tau is hyperphos-phorylated and has decreased solubility (Yoshiyama et al., 2007). When received, samples had been sectioned into cerebral cortex, hippocampus, cerebellum, brainstem, spinal cord and olfactory bulb and frozen at −80°C.
Western Blotting
For analysis of protein levels by Western blotting, cells were lysed in M-Per Protein Extraction Reagent including Halt Protease Inhibitor Cocktail and Halt Phosphatase Inhibitor Cocktail (Pierce, Rockford, IL). Insoluble cellular debris was removed by centrifugation at 12,000g for 15 min. Total protein concentration in cleared lysates was quantified using the BCA assay (Pierce) with bovine serum albumin (BSA) as a standard. Equal amounts of protein for each sample (typically 6.5 µg) were run on 4–20% SDS gels for 2 hr at 120 V. Gels were then transferred to nitrocellulose overnight (>16 hr) at 10 V. Nitrocellulose membranes were blocked for >2 hr with a blocking solution of either powdered milk or BSA in Tris-buffered saline with 0.1% Tween (TBST). Membranes were then incubated with primary antibody overnight at 4°C. The specific supplier, blocking conditions, incubation conditions, and secondary antibody for each primary antibody used in this work are listed in Table I. After three 10-min washes with TBST, membranes were incubated with an HRP-conjugated secondary antibody (GE Healthcare), and the blots were visualized with the ECL-Plus kit (GE Healthcare) on a Typoon 9400 scanner and quantified in ImageQuant software (GE Healthcare).
TABLE I.
Resources and Conditions for Western Blotting
| Antigen | Supplier | Catalogue no. | Dilution | Secondary antibody |
Blocking solution |
Antibody incubation solution |
|---|---|---|---|---|---|---|
| eIF2α | Biosource | AH01182 | 1:500 | Rabbit | 5% Milk | 1% BSA |
| Phos eIF2α | Invitrogen | 44728G | 1:1,000 | Rabbit | 5% BSA | 1% BSA |
| GAPDH | Invitrogen | 39–8600 | 1:15,000 | Mouse | 5% Milk | 1% BSA |
| GSK-3β | Cell Signaling | 9315 | 1:1,000 | Rabbit | 5% Milk | 5% BSA |
| Phos GSK-3β | Cell Signaling | 9336 | 1:500 | Rabbit | 5% Milk | 5% BSA |
| Tau p262 | Invitrogen | 44750G | 1:500 | Rabbit | 5% BSA | 3% BSA |
| Tau p396 | Invitrogen | 355300 | 1:500 | Mouse | 5% Milk | 3% Milk |
| Tau p199/202 | Invitrogen | 44768G | 1:1,000 | Rabbit | 5% BSA | 3% BSA |
| Tau 5 | Biosource | AHB0042 | 1:1,000 | Mouse | 5% Milk | 3% BSA |
Tau Solubility
The solubility of tau in cell culture samples was determined by sequential extraction using a series of solvents (Ishihara et al., 2001). Cells were lysed in high-salt reassembly buffer (RAB; 0.1 M MES, 0.5 mM MgSO4, 0.75 M NaCl and 0.02 M NaF at pH 7.0) to extract soluble as well as microtubule-bound tau. Insoluble material was separated from the RAB fraction by centrifugation at 13,000g for 30 min at 4°C. The resulting pellet was resuspended in the detergent containing-RIPA buffer (50 mM Tris-HCl, 150 mM NaCl2, 1% NP-40, 15 mM EDTA, 0.5% sodium deoxycholate, and 0.1% SDS at a pH of 8.0) to solubilize small tau aggregates. Insoluble material was separated from the RIPA fraction by centrifugation at 13,000g for 30 min at 4°C. The resulting pellet was resuspended in 70% formic acid to solubilize any remaining tau, including large aggregate species. The formic acid fraction was diluted 10× with 2 M Tris (pH 8.3) to reduce the acidity of the samples prior to analysis. Fractions were assayed for the presence of tau using Western blotting with the tau-5 antibody (Biosource).
RNA Analysis
Total RNA was purified from cultured cell samples using the SV Total RNA Isolation kit (Promega, Madison, WI) according to the manufacturer’s protocol. For brain tissue samples, tissue was homogenized using a PowerGen homogenizer (Fisher, Fair Lawn, NJ), and total RNA was isolated using Trizol reagent (Invitrogen), also according to the manufacturer’s instructions. Total RNA yields were measured by A260 using a NanoVue spectrophotometer (GE Healthcare).
Synthesis of cDNA from isolated RNA was performed with the ImProm-II Reverse Transcription System (Promega) according to the manufacturer’s instructions and with an oligo(dT)15 primer. After the reverse transcription reaction (RT-PCR), the reverse transcriptase was inactivated by incubation at 70°C for 15 min.
To determine the extent of XBP1 splicing, a segment of the XBP1 gene containing the region spliced by IRE1 during UPR activation was PCR amplified from cDNA. The primers used were mouse 5′-TTACGGGAGAAAACTCACGGA-3′ and 5′-GGGTCCAACTTGTCCAGAATGC-3′ and human 5′-ACGAGAGAAAACTCATGGC-3′ and 5′-GGGTCCAAGTTGTCCAGAATGC-3′. PCR conditions were 95°C for 5 min; 35 cycles of 95°C for 1 min, 49°C for 30 sec, and 72°C for 30 sec; and a final incubation at 72°C for 5 min. PCR products were separated on 2.5% agarose gels containing 0.00013% ethidium bromide for 2 hr at 100 V to visualize spliced and unspliced XBP1 fragments.
To determine levels of BiP and calreticulin (CRT) mRNA, quantitative PCR (qPCR) was performed on an ABI Prism 7000 Real-Time PCR instrument (Applied Biosystems, Foster City, CA) using DyNAmo HS SYBR Green dye (New England Biolabs, Boston, MA). Rpl 19, a ribosomal protein not regulated by ER stress (Lin et al., 2007), was used as an internal normalization standard. Primers used were mouse Rpl 19 5′-CTGATCAAGGATGGGCTGAT-3′ and 5′-TCAATCTTCTTGGATTCCCG-3′, mouse BiP 5′-GTCTGCTTCGTGTCTCCTCC-3′ and 5′-GGAATAGGTGGTCCCCAAGT-3′, and mouse calreticulin 5′-AAGAGCAGTTCTTGGACGGA-3′ and 5′-CACCAGTGTCTGGCCCTTAT-3′. PCR conditions were 95°C for 10 min, followed by 45 cycles of 95°C for 15 sec, 55°C for 30 sec, and 72°C for 30 sec. Each 30-µl qPCR included 1 µl of reverse transcription reaction and a primer concentration of 0.33 µM.
Quantification and Statistical Analysis of Gels
Western blots and DNA gels were quantified in Image-Quant software, and statistical analysis was performed in Minitab software. A one-way ANOVA test with a 95% confidence level was used to determine whether a statistically significant difference existed among the samples. In cases when a difference was found, Dunnett’s method (Dunnett, 1955) was used to identify samples that were significantly different from the DMSO-treated control.
RESULTS
Chemical Induction of UPR in SH-SY5Y Cells Did Not Affect Tau Biology
The effect of endoplasmic reticulum (ER) stress on tau biology was examined using SH-SY5Y cells as a model neuronal system (Biedler et al., 1978). Previous studies on SH-SY5Y cells have shown that addition of Ab oligomers can cause tau aggregation, but the mechanism of action was not explored (Ferrari et al., 2003). For the following experiments, cells were differentiated with retinoic acid to promote neuron-like morphology for at least 8 days prior to induction of UPR. The effect of cell differentiation on tau in SH-SY5Y cells has been well described in the literature. In undifferentiated cells, tau is localized predominantly in the nucleus and tau does not bind to microtubules in situ or in vitro (Tanaka et al., 1995; Uberti et al., 1997; Zhong et al., 1999). After differentiation, tau is localized primarily in the cytosol and along neuritic processes (Uberti et al., 1997). Differentiation has also been shown to change the pattern of expression of different tau isoforms (Uberti et al., 1997) and the phosphorylation of tau (Jamsa et al., 2004).
The UPR was induced in SH-SY5Y cells by treatment with either tunicamycin (TM), which inhibits N-linked glycosylation (Heifetz et al., 1979), or thapsigargin (TG), which disrupts calcium homeostasis (Thastrup et al., 1990; Sagara and Inesi, 1991). These chemical treatments have been used in many ER stress studies and are known to activate the UPR (Lin et al., 2007; Gonzales et al., 2008; Cho et al., 2009; Kim et al., 2009; Zhang et al., 2009). Cells were also treated with okadaic acid (OA), a phosphatase inhibitor, as a positive control for tau phosphorylation (Perez et al., 2002; Xu et al., 2004).
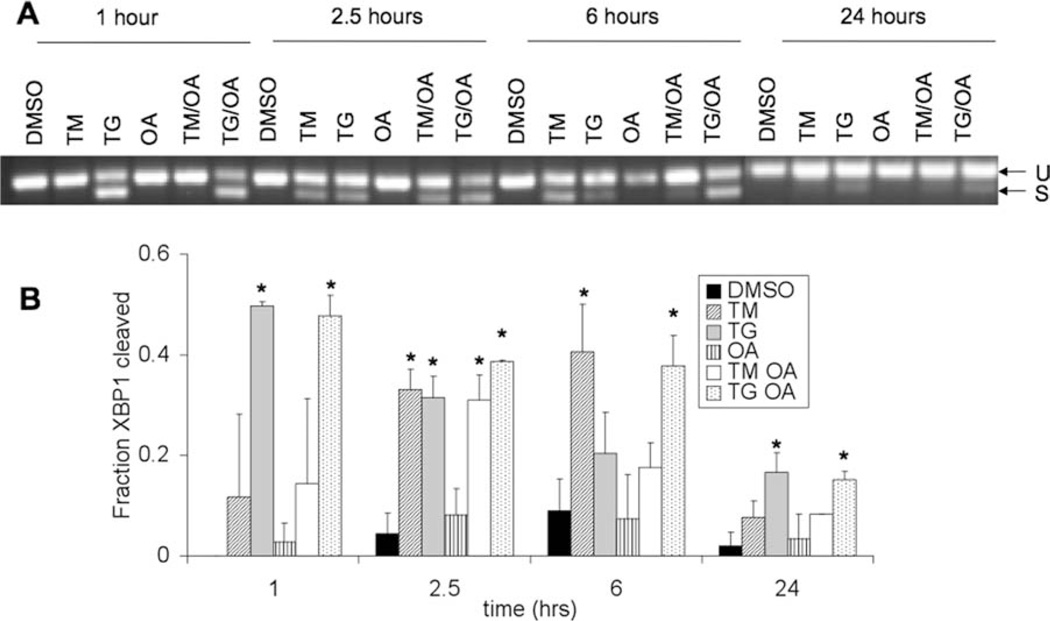
To confirm that the UPR was activated in our experimental system, we monitored several UPR markers following addition of TM, TG, and OA. Results for splicing of XBP1 mRNA (an early event in the IRE1 branch of the UPR) are shown in Figure 1. As expected, treatment with TM or TG caused splicing of XBP1 beginning at 1 hr for TG and 2.5 hr for TM and for times up to 6 hr, indicating that the IRE1 branch of the UPR is activated in these samples. Attenuation of splicing was seen at 24 hr, as expected (Lin et al., 2007). The percentage of spliced XBP1 was determined from quantification of band intensities in the gel, and results with statistically significant levels of splicing (compared with DMSO control) are indicated in Figure 1B. No significant XBP1 splicing was observed in cells treated with OA alone.
Fig. 1.
SH-SY5Y cells, 8 days following differentiation by retinoic acid, were untreated (DMSO) or treated with tunicamycin (TM), thapsigargin (TG), okadaic acid (OA), or a combination of OA and TM or TG as described in Materials and Methods. A: A segment of the XBP1 gene containing the region spliced by IRE1 was amplified from cDNA of cell lysates by PCR. PCR products were separated on a 2.5% agarose gel to resolve unspliced (U) and spliced (S) XBP1. B: Quantification of XBP1 splicing from the agarose gels was performed as described in Materials and Methods. Error bars represent the standard deviation from the mean of two biological replicates. Asterisks indicate a statistically significant difference from DMSO control with a 95% confidence level.
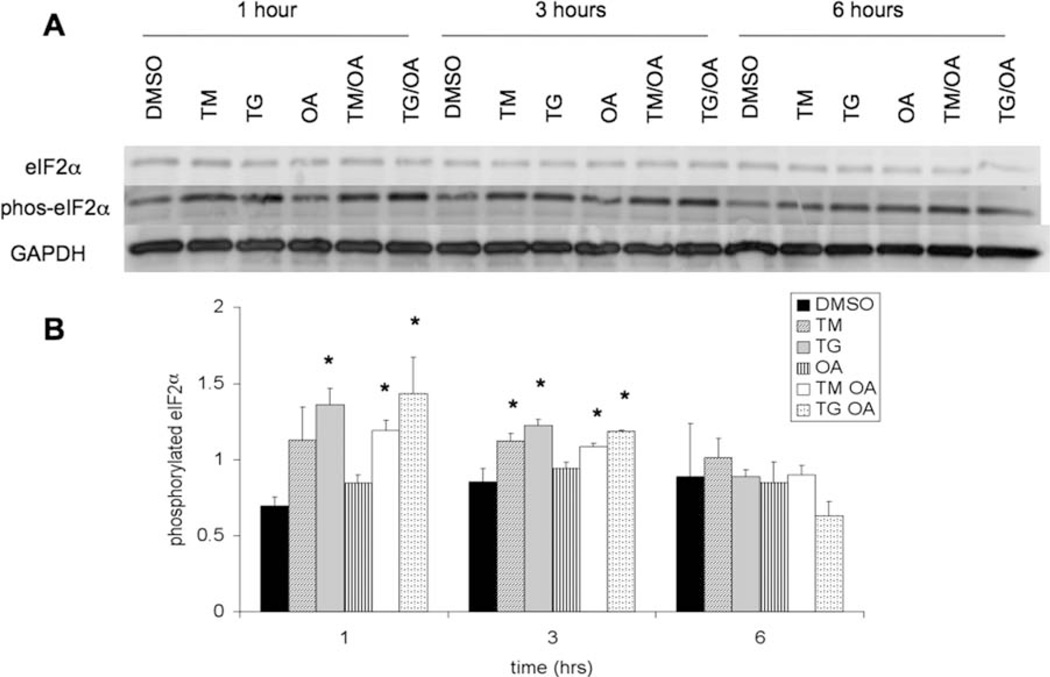
Phosphorylation of eIF2α (an early event in the PERK branch of the UPR) was also monitored. Western blots of total eIF2α levels and phosphorylated eIF2α levels are shown in Figure 2A. Phosphorylation of eIF2α was higher 1 and 3 hr after treatment with TM or TG, indicating that the PERK branch of the UPR is activated in these cases. Increased eIF2α phosphorylation was not observed in samples treated with OA alone. Figure 2B shows quantification of phosphorylated eIF2α levels, and statistically significant differences from DMSO control are indicated. Total eIF2α levels were also quantified, and, as expected, no statistically significant differences were observed among samples.
Fig. 2.
A: Levels of total eIF2α and phosphorylated eIF2α in lysates from SH-SY5Y cells, which were treated as described in Figure 1, were measured by Western blotting. Glyceraldehyde-3-phosphate de-hydrogenase (GAPDH) was used as a protein loading control. B: Quantification from Western analysis of phosphorylated eIF2α normalized to GAPDH levels. Error bars represent the standard deviation from the mean of two biological replicates. Asterisks indicate a statistically significant difference from DMSO control with a 95% confidence level.
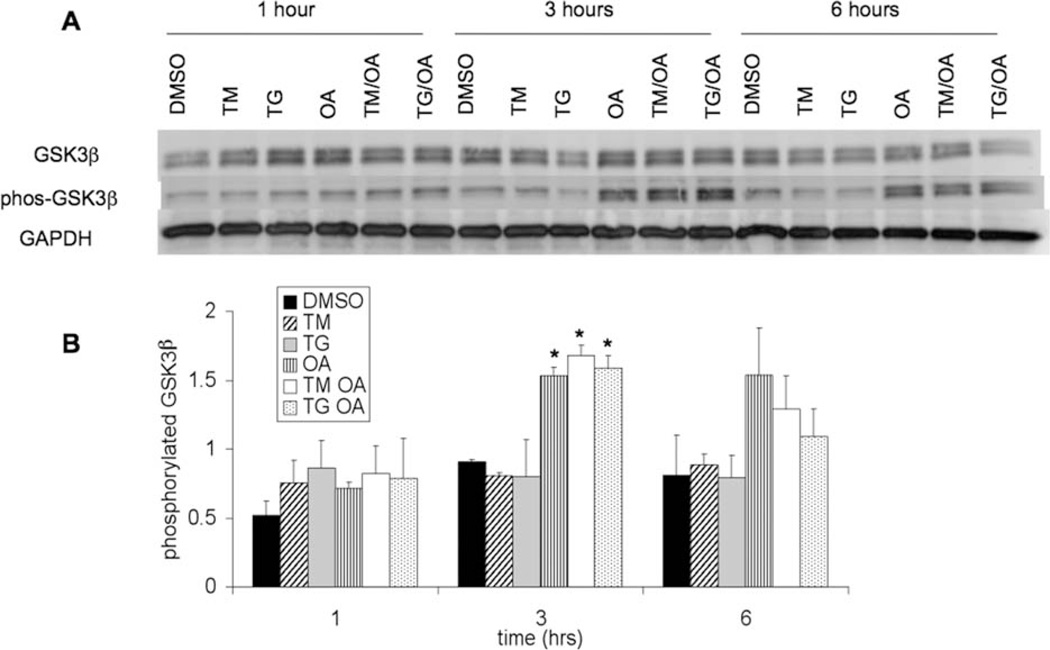
It has been reported that the activity of GSK-3β, a known tau kinase, is increased by the UPR (Song et al., 2002; Brewster et al., 2006). Because this provides a potential mechanistic link between the UPR and tau biology, we examined GSK-3β levels in TM-, TG-, and OA-treated samples. Figure 3A shows Western blots of total GSK-3β levels and GSK-3β phosphorylated at serine 9. Phosphorylation of GSK-3β at this site inhibits kinase activity; thus, an increase in phosphorylation would decrease activity. Quantification of the phosphorylated GSK-3β levels is shown in Figure 3B. In the blot shown, a small decrease in GSK-3β phosphorylation was observed in samples treated with TM at 6 hr and samples treated with TG at 3 and 6 hr. This difference was not apparent in the replicate blot, and, when quantified and averaged, the change was not significant. This indicates that GSK-3β was not significantly activated by the UPR in this system. Samples treated with OA showed increased phosphorylation of GSK-3β at 3 and 6 hr resulting from nonspecific phosphatase inhibition, as expected.
Fig. 3.
A: Levels of total GSK-3β and phosphorylated GSK-3β in lysates from SH-SY5Y cells, which were treated as described in Figure 1, were measured by Western blotting. GAPDH was used as a protein loading control. B: Quantification from Western analysis of phosphorylated GSK-3β levels normalized to GAPDH levels. Error bars represent the standard deviation from the mean of two biological replicates. Asterisks indicate a statistically significant difference from DMSO control with 95% confidence level.
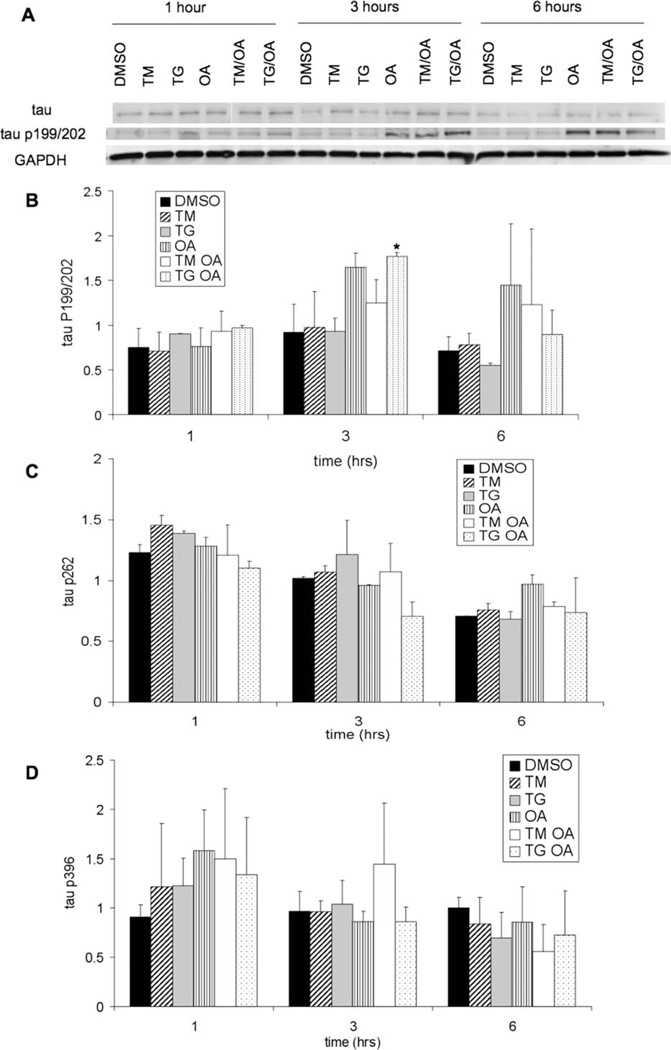
To determine what effect UPR activation has on tau biology, tau solubility and extent of phosphorylation were tested. Samples of cells treated with TM, TG, and OA were analyzed by Western blotting using tau-5 antibody, which recognizes all tau species, and phosphospecific antibodies, which recognize only tau that is phos-phorylated at specific sites. Representative Western blots of total tau and tau phosphorylated at serine 199 or 202 are shown in Figure 4A. Quantifications of Western blots for phosphorylated tau species are shown in Figure 4B (serine 199/202), Figure 4C (serine 262), and Figure 4D (serine 396). Hyperphosphorylation of tau at these sites is associated with increased propensity to form tangles (Buee et al., 2000). Treatment with OA, a phophatase inhibitor, caused an increase in levels of tau phosphorylation at serine 199/202, as previously reported (Perez et al., 2002), but no significant increase in phosphorylation of tau at serine 262 or serine 396. However, this is not surprising, insofar as large variability in OA-induced phosphorylation among different tau sites has been observed previously. A recent study in cultured neurons reported a 450% increase in phosphorylation at serine 199/ 202 but only a 20% increase in phosphorylation at serine 396 upon treatment with 100 nM OA. Interestingly, samples treated with only TM or TG did not show any significant change in tau phosphorylation levels (Rahman et al., 2009). Total tau levels were also quantified, and no statistically significant differences existed among samples. When tau solubility was assayed by sequential extraction using RAB, RIPA, and 70% formic acid, tau was observed only in the RAB fraction (data not shown), indicating that no insoluble aggregates were formed.
Fig. 4.
A: Levels of total tau and tau phosphorylated at serine 199 or 202 in lysates from SH-SY5Y cells, which were treated as described in Figure 1, were measured by Western blotting. GAPDH was used as a loading control. Quantification from Western analysis of phosphorylated tau levels normalized to GAPDH levels for phosphorylation at serine 199/202 (B), serine 262 (C), and serine 396 (D). Error bars represent the standard deviation from the average of two (B,C) or three (D) biological replicates. Asterisk indicates statistically significant difference from DMSO control with a 95% confidence level.
Overexpression of WT and P301L Tau Did Not Induce the UPR
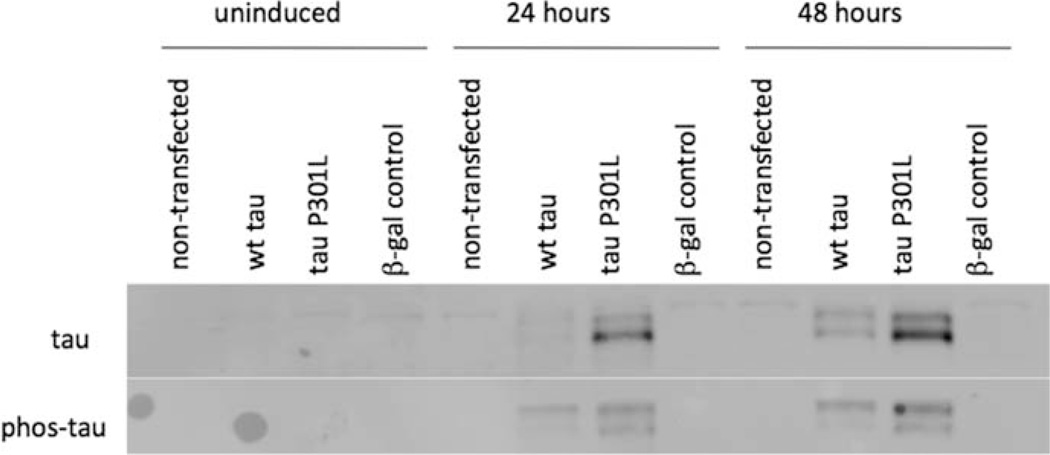
To determine whether tau dysfunction could be a cause of ER stress, both wild-type (WT) tau and a mutant form of tau (P301L), along with a control protein, β-galactosidase (β-gal), were overexpressed in HEK293 cells. The commercial tetracycline-regulated T-REx expression system (Invitrogen) was used to allow control of tau expression. Figure 5 shows Western blots of tau levels in uninduced cells and cells 24 and 48 hr after protein expression was induced with 1 lg/ml tetracycline. Tau was observed in the WT and P301L samples following induction but not in the uninduced samples or the nontransfected and β-gal control samples, confirming that these cell lines express tau only under control of the tetracycline-induced promoter. Two tau bands are observed on the Western blot. This has been previously observed when tau was recombinantly expressed in CHO cells, and the difference in mobility was attributed to different degrees of phosphorylation (Vogelsberg-Ragaglia et al., 2000). The increased expression level of the P301L tau compared with that of WT tau most likely was due to stochastic differences in transfection efficiency. Previous work using this system to express human transferrin has shown that the transfection generates expression clones with a wide range of expression levels (Jones et al., 2005).
Fig. 5.
T-REx 293 cells, which were either untransfected or stably transfected with a plasmid containing the gene for either WT tau, P301L tau, or β-galactosidase (β-gal), were treated with tetracycline to induce protein expression as described in Materials and Methods. Expression of recombinant tau was confirmed by Western blotting for tau and phosphorylated tau (serines 199/202).
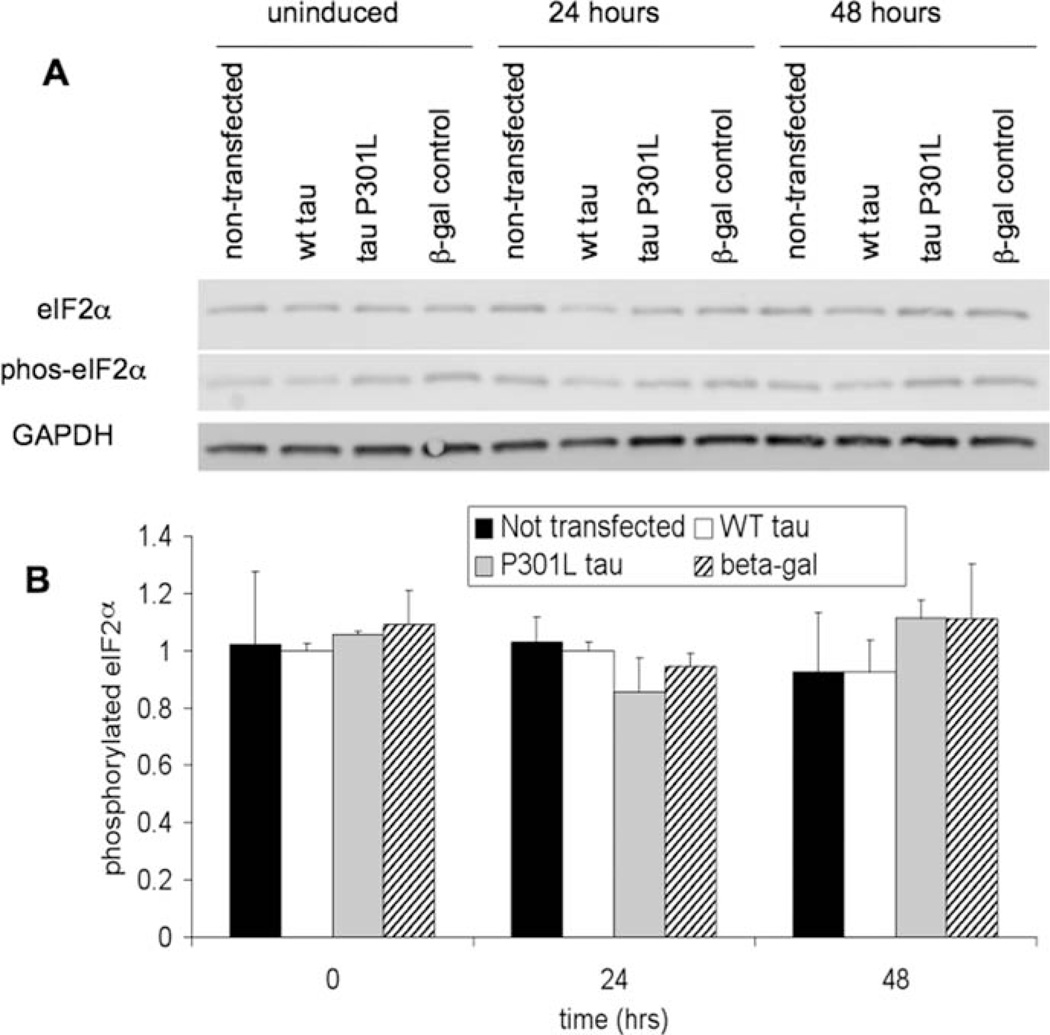
Once expression of tau was confirmed, cell samples were tested for UPR activation. Figure 6 shows splicing of XBP1 mRNA with a positive control from treatment of SH-SY5Y cells with TG for 1 hr. None of the samples show any splicing of XBP1, indicating that the IRE1 branch of the UPR is not activated by WT or P301L tau expression in T-REx 293 cells. Figure 7A shows a representative Western blot of total and phosphorylated eIF2α in these samples, and quantification of phosphorylated eIF2α levels is shown in Figure 7B. No statistically significant changes in total or phosphorylated eIF2α levels were observed, indicating the PERK branch of the UPR was not activated by WT or P301L expression in T-REx 293 cells. Taken together, the XBP1 and eIF2α results indicate that overexpression of WT or P301L tau in this system did not activate the UPR.
Fig. 6.
Protein expression was induced in T-REx 293 cells as described for Figure 5. A segment of the XBP1 gene containing the region spliced by Ire1 was amplified via PCR from cDNA collected from T-REx 293 cell lysates. PCR products were separated on a 2.5% agarose gel to resolve unspliced (U) and spliced (S) XBP1. SH-SY5Y cells treated with TG for 1 hr were used as a positive control. Results shown are representative of two biological replicates.
Fig. 7.
Protein expression was induced in T-REx 293 cells as described in Figure 5. A: Levels of total eIF2α and phosphorylated eIF2α in T-REx 293 cell lysates were measured by Western blotting. GAPDH was used as a protein loading control. B: Quantification from Western analysis of phosphorylated eIF2α levels normalized to GAPDH levels. Error bars represent the standard deviation from the average of two biological replicates.
The UPR Was Not Induced in PS19 Transgenic Mice
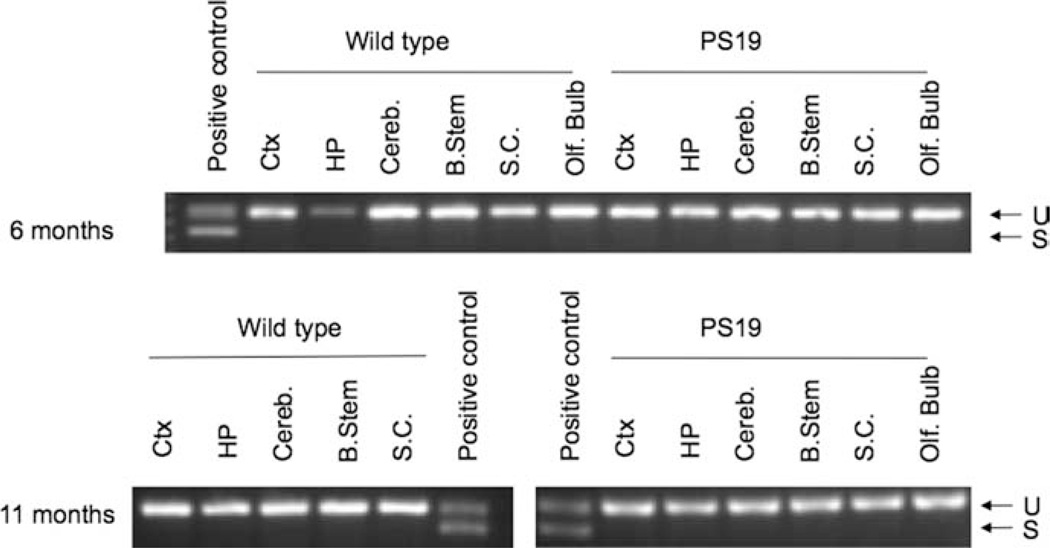
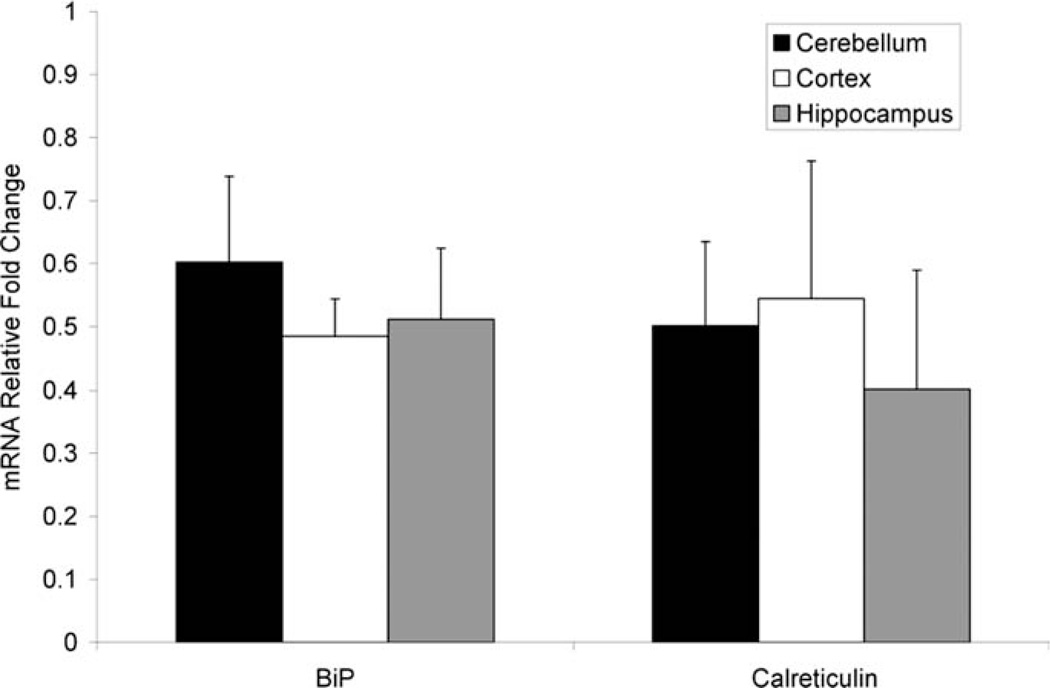
Although the UPR was not observed in T-REx 293 cells expressing P301L or WT tau, transient overex-pression of tau in a cell culture system is significantly different from the physiological progression of tauopathies, in which tau accumulates and aggregates over long periods of time. To test further for the ability of tau dysfunction to induce the UPR, brain tissue samples from transgenic mice expressing P301S tau were tested for XBP1 splicing. PS19 mice have mild neurological pathology at 6 months and severe pathology at 11 months (Yoshiyama et al., 2007). Figure 8 shows representative XBP1 splicing results for both WT and PS19 transgenic mice at 6 and 11 months of age. XBP1 splicing was not observed in any of the samples, indicating that the UPR was not activated in this transgenic mouse model. In addition, the levels of BiP and calreticulin mRNA were measured by qPCR in samples from mice at 11 months of age. The relative -fold increase in mRNA levels for PS19 mice compared with a nontransgenic control are plotted in Figure 9. BiP and calreticulin are both ER-resident chaperones that are up-regulated during ER stress. BiP mRNA levels have been observed to increase more than tenfold upon treatment of cultured cells with TM and more than twofold during protein overexpression (Lin et al., 2007). Calreticulin mRNA levels have been reported to increase more than twofold upon during TM treatment of Caenorhabditis elegans (Lee et al., 2007), and increased expression of the calreticulin gene has been observed in cultured cells treated with TG (Waser et al., 1997). Interestingly, in the tissue samples from PS19 mice examined here, BiP and calreticulin mRNA levels were lower than levels in the nontransgenic control. The cause of this down-regulation of BiP and calreticulin is not known. However, BiP and calreticulin were clearly not up-regulated, providing further conformation that the UPR was not activated in the PS19 transgenic mice.
Fig. 8.
A segment of the XBP1 gene containing the region spliced by Ire1 was amplified via PCR from cDNA of WT and PS19 mouse tissue samples. PCR products were separated on a 2.5% agarose gel to resolve unspliced (U) and spliced (S) XBP1. SH-SY5Y cells treated with TM for 2.5 hr (6 months and 11 months PS19) or TG for 1 hr (11 months WT) were used as a positive control. Results shown for PS19 mice are representative of three replicate specimens. Ctx, cerebral cortex; HP, hippocampus; Cereb, cerebellum; B. Stem, brainstem; S.C., spinal cord; Olf. Bulb, olfactory bulb.
Fig. 9.
Levels of BiP and calreticulin mRNA in tissue samples from the cerebral cortex, hippocampus, and cerebellum were measured by qPCR and normalized to levels of Rpl19, a ribosomal protein not regulated by ER stress. The relative -fold changes in mRNA levels for PS19 mice compared with a nontransgenic control are plotted. Error bars represent standard deviation from the mean of samples from three transgenic animals.
DISCUSSION
ER stress and activation of the UPR have been implicated in several diseases, including Alzheimer’s disease, although the role of the stress response in disease pathologies is not known (Lindholm et al., 2006; Yoshida, 2007; Scheper and Hoozemans, 2009). ER stress has been observed in brain tissue of Alzheimer’s disease patients (Hoozemans et al., 2005, 2009) and in combination with tau aggregation when cultured neurons are treated with Aβ oligomers (Resende et al., 2008). However, it is unclear whether formation of tau aggregates induces ER stress or whether activation of the UPR contributes to tau aggregation. It is also possible that tau aggregation and ER stress are separate events and are not directly linked. This work tests the relationship between tau biology and the UPR in several model systems in an attempt to find a direct, mechanistic link between the two. Surprisingly, no correlation between UPR activation and tau was observed in any of the systems tested.
The first model system described here was used to test the hypothesis that activation of the UPR could cause changes in tau biology such as increased phospho-rylation and propensity to form tangles. This hypothesis is consistent with recent experimental results from other groups. In a study by Resende and colleagues (2008), cultured neuronal cells were treated with Aβ oligomers, resulting in hyperphosphorylation of tau, activation of GSK-3β, and increases in BiP levels. Treatment of cells with a selective inhibitor of GSK-3β or dantroline, an inhibitor of ER calcium release, decreased the extent of Aβ-induced phosphorylation of tau, indicating that GSK-3β activation and ER stress were involved in tau phosphorylation in this system. The authors propose that Aβ oligomers cause ER stress, which in turn activates GSK-3β, leading to increased phosphorylation of tau. A significant unanswered question in this study is how extracellular Aβ oligomers trigger ER stress. However, if Aβ effects on tau are the result of ER stress, then induction of ER stress by other methods should result in a similar sequence of events. The results described here indicate that this is not the case.
In our studies, ER stress was induced by two different chemical treatments, thapsigargin and tunicamycin, and activation of the UPR was confirmed by the UPR signaling markers XBP1 and eIF2α. However, no changes in tau phosphorylation or solubility were observed. In addition, activation of GSK-3β was not conclusively demonstrated. It is possible that ER stress is one of multiple results of Aβ treatment. For example, there is evidence that Aβ addition causes inhibition of the proteosome (Oddo, 2008). The combination of ER stress and other factors may be necessary to cause tau hyperphosphorylation and aggregation. It is also possible that ER stress is a result of Aβ treatment but does not play a causative role in tau hyperphosphorylation. Although the mechanism by which Aβ increases tau phosphorylation remains unclear, the results presented here indicate that UPR activation alone is not sufficient to lead to hyperphosphorylation of tau.
One limitation of these experiments is that the chemical reagents used cause acute ER stress, which causes sustained UPR activation and eventual apoptosis. Under conditions of low or moderate stress, the UPR can result in a return to cellular homeostasis. The specific factors that determine the balance between proapoptotic and prosurvival responses to ER stress and the effects of prolonged or repeated low-level ER stress are not yet well understood (Rutkowski et al., 2006; Lin et al., 2007). However, chronic, low-level stress may play a significant role in neurodegeneration and other protein folding disorders and warrants increased study in the future.
The second hypothesis examined here is that tau dysfunction could lead to ER stress and UPR activation. To test this hypothesis, an experimental system that models aggregation-prone tau states was needed. Researchers have identified at least 37 mutations in the tau gene that are associated with frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) and other, similar disorders (Wolfe, 2009). To mimic the disease state, two of the FTDP-17 variants of tau (P301L and P301S) were used. The P301L variant was recombinantly overexpressed in cultured cells, and the P301S variant was expressed in transgenic mice. In both of these model systems, no evidence of ER stress or UPR activation was observed, indicating that these disease-like tau variants do not lead to ER stress. This result is particularly convincing in the case of the trans-genic mouse model, for which tau is known to aggregate and cause neurological impairment.
The inability of tau to induce the UPR in the model systems described here in combination with previous work demonstrating that extracellular Aβ alone can cause UPR induction in cultured neurons indicates that Aβ is the most likely cause of the ER stress that is observed in AD patients. However, ER stress has also been observed in other neurodegenerative disorders (Scheper and Hoozemans, 2009), including Parkinson’s disease (Hoozemans et al., 2007) and amyotrophic lateral sclerosis (Atkin et al., 2008), indicating that activation of this response is not unique to aggregation of Aβ. This is the first report of which the authors are aware that suggests no link between tau aggregation and ER stress in neurodegenerative disorders.
ACKNOWLEDGMENTS
The authors thank Dr. Virginia Lee (University of Pennsylvania) for supplying tissue samples; Dr. Jeffery Twiss, Dr. Carolyn Schanen, and Hunter Stitik (UD Biological Sciences) for technical help with tissue samples; Dr. Patrick Daugherty (UCSB) for generous use of laboratory facilities; and Dr. Kenneth Kosik (UCSB) for the tau 4R gene.
Contract grant sponsor: National Institutes of Health; Contract grant number: F33 AG031610 (to A.S.R.); Contract grant number: R01 GM075297; Contract grant number: P20 RR015588; Contract grant sponsor: National Science Foundation; Contract grant number: DGE-0221651 (to M.L.S.).
REFERENCES
- Alzheimer’s Association. 2009 Alzheimer’s disease facts and figures. Alzheimers Dementia. 2009;5 doi: 10.1016/j.jalz.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neuro-biol Dis. 2008;30:400–407. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38:3751–3757. [PubMed] [Google Scholar]
- Brandt R, Hundelt M, Shahani N. Tau alteration and neuronal degeneration in tauopathies: mechanisms and models. Biochim Biophys Acta. 2005;1739:331–354. doi: 10.1016/j.bbadis.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Brewster JL, Linseman DA, Bouchard RJ, Loucks FA, Precht TA, Esch EA, Heidenreich KA. Endoplasmic reticulum stress and trophic factor withdrawal activate distinct signaling cascades that induce glyco-gen synthase kinase-3 beta and a caspase-9-dependent apoptosis in cere-bellar granule neurons. Mol Cell Neurosci. 2006;32:242–253. doi: 10.1016/j.mcn.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- Chafekar SM, Hoozemans JJ, Zwart R, Baas F, Scheper W. Abeta 1–42 induces mild endoplasmic reticulum stress in an aggregation state-dependent manner. Antioxid Redox Signal. 2007;9:2245–2254. doi: 10.1089/ars.2007.1797. [DOI] [PubMed] [Google Scholar]
- Cho YM, Jang YS, Jang YM, Chung SM, Kim HS, Lee JH, Jeong SW, Kim IK, Kim JJ, Kim KS, Kwon OJ. Induction of unfolded protein response during neuronal induction of rat bone marrow stromal cells and mouse embryonic stem cells. Exp Mol Med. 2009;41:440–452. doi: 10.3858/emm.2009.41.6.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudna RE, Dickson AJ. Endoplasmic reticulum signaling as a determinant of recombinant protein expression. Biotechnol Bioeng. 2003;81:56–65. doi: 10.1002/bit.10445. [DOI] [PubMed] [Google Scholar]
- Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096–1121. [Google Scholar]
- Ferrari A, Hoerndli F, Baechi T, Nitsch RM, Gotz J. Beta-amyloid induces paired helical filament-like tau filaments in tissue culture. J Biol Chem. 2003;278:40162–40168. doi: 10.1074/jbc.M308243200. [DOI] [PubMed] [Google Scholar]
- Gonzales JC, Gentile CL, Pfaffenbach KT, Wei Y, Wang D, Pagliassotti MJ. Chemical induction of the unfolded protein response in the liver increases glucose production and is activated during insulin-induced hypoglycaemia in rats. Diabetologia. 2008;51:1920–1929. doi: 10.1007/s00125-008-1094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tuni-camycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phos-phate transferase. Biochemistry. 1979;18:2186–2192. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, Eikelenboom P, Scheper W. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol. 2005;110:165–172. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemul-ler JM, Scheper W. Activation of the unfolded protein response in Parkinson’s disease. Biochem Biophys Res Commun. 2007;354:707–711. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelen-boom P, Scheper W. The unfolded protein response is activated in pretangle neurons in alzheimer’s disease hippocampus. Am J Pathol. 2009;174:1241–1251. doi: 10.2353/ajpath.2009.080814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Ad-amson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snow-den J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Higuchi M, Zhang B, Yoshiyama Y, Hong M, Trojanowski JQ, Lee VMY. Attenuated neurodegenerative disease phenotype in tau transgenic mouse lacking neurofilaments. J Neurosci. 2001;21:6026–6035. doi: 10.1523/JNEUROSCI.21-16-06026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsa A, Hasslund K, Cowburn RF, Backstrom A, Vasange M. The retinoic acid and brain-derived neurotrophic factor differentiated SH-SY5Y cell line as a model for Alzheimer’s disease-like tau phospho-rylation. Biochem Biophys Res Commun. 2004;319:993–1000. doi: 10.1016/j.bbrc.2004.05.075. [DOI] [PubMed] [Google Scholar]
- Jones J, Nivitchanyong T, Giblin C, Ciccarone V, Judd D, Gorfien S, Krag SS, Betenbaugh MJ. Optimization of tetracycline-responsive recombinant protein production and effect on cell growth and ER stress in mammalian cells. Biotechnol Bioeng. 2005;91:722–732. doi: 10.1002/bit.20566. [DOI] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Manabe T, Hitomi J, Kudo T, Tohyama M. Induction of neuronal death by ER stress in Alzheimer’s disease. J Chem Neuroanat. 2004;28:67–78. doi: 10.1016/j.jchemneu.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kim I, Shu CW, Xu W, Shiau CW, Grant D, Vasile S, Cosford ND, Reed JC. Chemical biology investigation of cell death pathways activated by endoplasmic reticulum stress reveals cytoprotective modulators of ASK1. J Biol Chem. 2009;284:1593–1603. doi: 10.1074/jbc.M807308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, Teodoro T, Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology. 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- Lee D, Singaravelu G, Park BJ, Ahnn J. Differential requirement of unfolded protein response pathway for calreticulin expression in Cae-norhabditis elegans. J Mol Biol. 2007;372:331–340. doi: 10.1016/j.jmb.2007.06.071. [DOI] [PubMed] [Google Scholar]
- Lee VM. Biomedicine. Tauists and beta-aptists united—well almost! Science. 2001;293:1446–1447. doi: 10.1126/science.1064684. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegener-ative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo S. The ubiquitin-proteasome system in Alzheimer’s disease. J Cell Mol Med. 2008;12:363–373. doi: 10.1111/j.1582-4934.2008.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, Hernandez F, Gomez-Ramos A, Smith M, Perry G, Avila J. Formation of aberrant phosphotau fibrillar polymers in neural cultured cells. Eur J Biochem. 2002;269:1484–1489. doi: 10.1046/j.1432-1033.2002.02794.x. [DOI] [PubMed] [Google Scholar]
- Rahman A, Ting K, Cullen KM, Braidy N, Brew BJ, Guillemin GJ. The excitotoxin quinolinic acid induces tau phosphorylation in human neurons. PLoS ONE. 2009;4:e6344. doi: 10.1371/journal.pone.0006344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende R, Ferreiro E, Pereira C, Oliveira CR. ER stress is involved in Abeta-induced GSK-3beta activation and tau phosphoryla-tion. J Neurosci Res. 2008;86:2091–2099. doi: 10.1002/jnr.21648. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara Y, Inesi G. Inhibition of the sarcoplasmic reticulum Ca2+ transport ATPase by thapsigargin at subnanomolar concentrations. J Biol Chem. 1991;266:13503–13506. [PubMed] [Google Scholar]
- Scheper W, Hoozemans JJ. Endoplasmic reticulum protein quality control in neurodegenerative disease: the good, the bad and the therapy. Curr Med Chem. 2009;16:615–626. doi: 10.2174/092986709787458506. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Phys-iol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Song L, De Sarno P, Jope RS. Central role of glycogen synthase kinase-3beta in endoplasmic reticulum stress-induced caspase-3 activation. J Biol Chem. 2002;277:44701–44708. doi: 10.1074/jbc.M206047200. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Fitzgerald U, Samali A. Caspase-12 and ER-stress-mediated apoptosis: the story so far. Ann N Y Acad Sci. 2003;1010:186–194. doi: 10.1196/annals.1299.032. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Iqbal K, Trenkner E, Liu DJ, Grundke-Iqbal I. Abnormally phosphorylated tau in SY5Y human neuroblastoma cells. FEBS Lett. 1995;360:5–9. doi: 10.1016/0014-5793(95)00061-d. [DOI] [PubMed] [Google Scholar]
- Thastrup O, Cullen PJ, Drobak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2+-ATPase. Proc Natl Acad Sci U S A. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti D, Rizzini C, Spano PF, Memo M. Characterization of tau proteins in human neuroblastoma SH-SY5Y cell line. Neurosci Lett. 1997;235:149–153. doi: 10.1016/s0304-3940(97)00715-5. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A. Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiol Rev. 2005;85:201–279. doi: 10.1152/physrev.00004.2004. [DOI] [PubMed] [Google Scholar]
- Vogelsberg-Ragaglia V, Bruce J, Richter-Landsberg C, Zhang B, Hong M, Trojanowski JQ, Lee VM. Distinct FTDP-17 missense mutations in tau produce tau aggregates and other pathological phenotypes in transfected CHO cells. Mol Biol Cell. 2000;11:4093–4104. doi: 10.1091/mbc.11.12.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waser M, Mesaeli N, Spencer C, Michalak M. Regulation of cal-reticulin gene expression by calcium. J Cell Biol. 1997;138:547–557. doi: 10.1083/jcb.138.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS. Tau mutations in neurodegenerative diseases. J Biol Chem. 2009;284:6021–6025. doi: 10.1074/jbc.R800013200. [DOI] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Xu YF, Zhang YJ, Zhang AH, Zhang Q, Wu T, Wang JZ. Attenuation of okadaic acid-induced hyperphosphorylation of cytos-keletal proteins by heat preconditioning and its possible underlying mechanisms. Cell Stress Chaperones. 2004;9:304–312. doi: 10.1379/CSC-23R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Zhang LJ, Chen S, Wu P, Hu CS, Thorne RF, Luo CM, Hersey P, Zhang XD. Inhibition of MEK blocks GRP78 up-regulation and enhances apoptosis induced by ER stress in gastric cancer cells. Cancer Lett. 2009;274:40–46. doi: 10.1016/j.canlet.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Zhong J, Iqbal K, Grundke-Iqbal I. Hyperphosphorylated tau in SY5Y cells: similarities and dissimilarities to abnormally hyperphos-phorylated tau from Alzheimer disease brain. FEBS Lett. 1999;453:224–228. doi: 10.1016/s0014-5793(99)00715-2. [DOI] [PubMed] [Google Scholar]