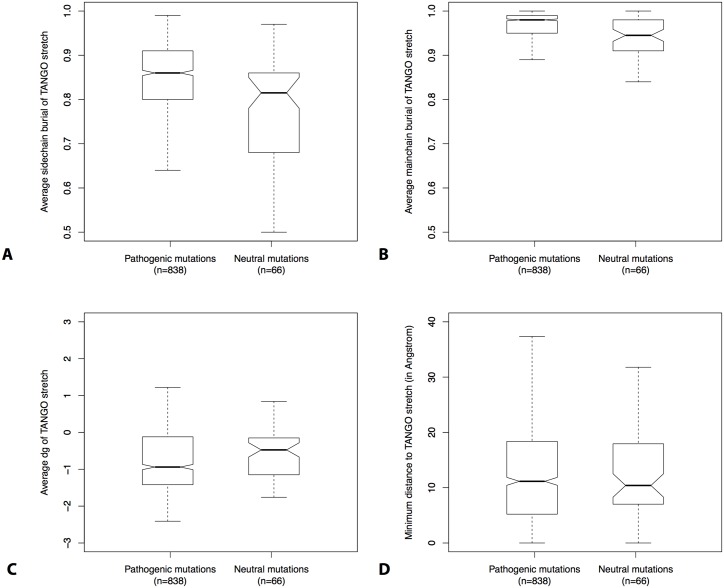
Fig 5. Structural information about APRs.
(A-C) Boxplot of the average A) sidechain burial, B) mainchain burial and C) stability (dG) of the APR calculated by FoldX. A negative dG indicates that the residue contributes to the thermodynamic stability of the protein. (D) The minimum distance in structural space between the mutation and the strongest APR present in the domain.