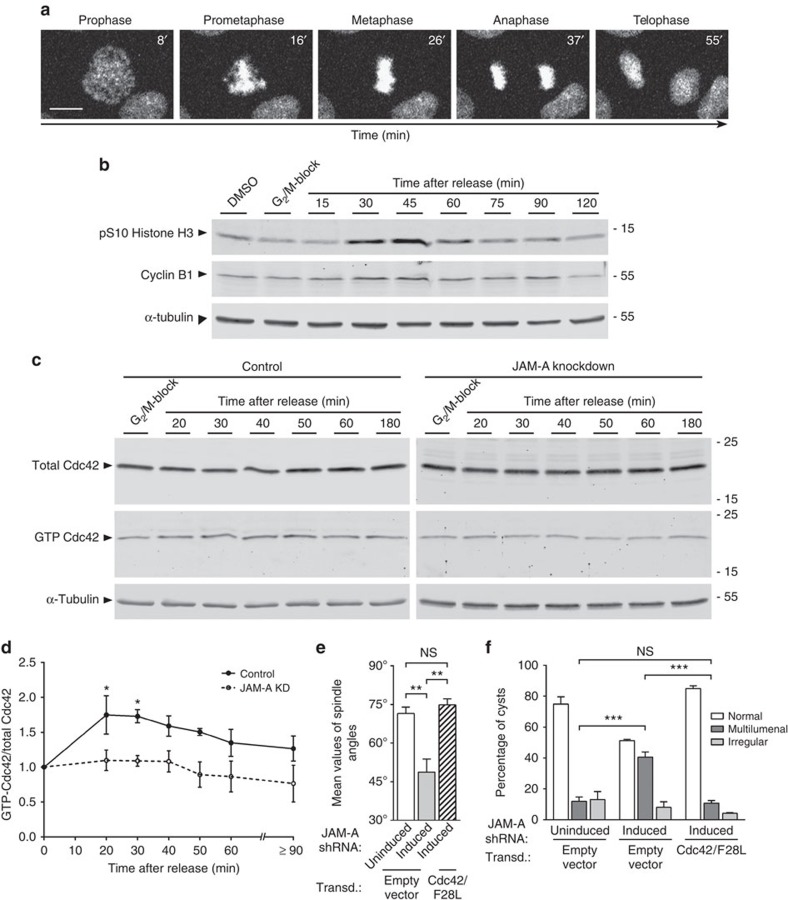
Figure 3. JAM-A controls Cdc42 activity during mitosis.
(a) Mitosis in MDCK cells. MDCK cells were stained with Hoechst 33342 and observed under a fluorescence microscope. Images representative for distinct stages of mitosis were taken at the indicated time points. Mitosis was typically completed within 60 min after the first signs of nuclear condensation. Size bar, 5 μm. (b) RO-3306-treated cells proceed normally through mitosis after release from the RO-3306 block. MDCK cells were treated with RO-3306 as described in the Methods section. Cells were harvested at the indicated time points and analysed for the levels of two markers of mitosis, Ser10-phosphorylated histone H3 and Cyclin B1. (c) JAM-A activates Cdc42 during mitosis. MDCK cells stably transfected with a doxycycline-regulated JAM-A shRNA expression vector were either left untreated (control) or were treated with doxycycline to induce JAM-A shRNA expression (JAM-A knockdown), then synchronized with RO-3306 to arrest cells at G2/M-phase transition (G2/M block) and analysed for Cdc42 activity at the indicated time points after release from the G2/M block. (d) Quantification of Cdc42 activity of five independent experiments. Levels of active Cdc42 (GTP-Cdc42) were normalized to total Cdc42 levels and are expressed as signal intensities relative to the normalized GTP-Cdc42 levels of cells that were not released into mitosis (G2/M block). *P<0.05. (e,f) Cdc42/F28L restores planar spindle orientation and single lumen specification in JAM-A knockdown cells. MDCK cells with doxycycline-inducible JAM-A knockdown were transduced with lentiviral vectors expressing Cdc42/F28L. Cells were grown to cysts in 3D collagen gels. Spindle orientation (e) and lumen formation (f) were analysed in three independent experiments as described in Figs 1 and 2, respectively. Statistical analysis was performed using two-way repeated-measures ANOVA with Bonferroni's post-hoc test (d) or one-way ANOVA with Tukey's post-hoc test (e,f). The rescue experiments using Cdc42/F28L to restore mitotic spindle orientation (e) and single lumen specification (f) were performed in parallel with the rescue experiments using mJAM-A (shown in Figs 1e and 2b, respectively); the control samples are therefore identical. Data are expressed as means±s.e.m.; ns, not significant; *P<0.05, **P<0.01, ***P<0.001.