Fig. 2.
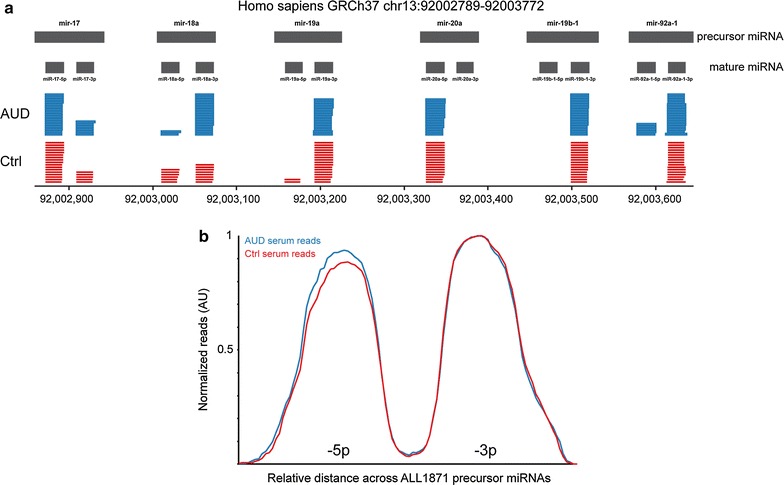
MicroRNAs in serum are in mature form. a Representative image showing RNA-seq reads aligned to region on chromosome 13. Reads are concentrated on mature miRNA chromosome coordinates as defined by the miRBase 20 database. b Trend plots mapping reads across all precursor miRNAs in the human genome show that reads are concentrated in regions that roughly correspond to mature miRNAs (-5p or -3p regions). We point out that the data shown are merely illustrative of the alignments produced during analysis of the RNA-Seq reads. Note that mature miRNAs are derived from the same strand as the full-length precursor, and appear in the location specified by the -5p box or -3p box. Also note that while the absolute levels of some mature miRNAs appeared to differ between AUD and Control subjects, the pattern of -5p or -3p usage appeared to be highly similar, which is best appreciated for the entire set of data in the lower plot
