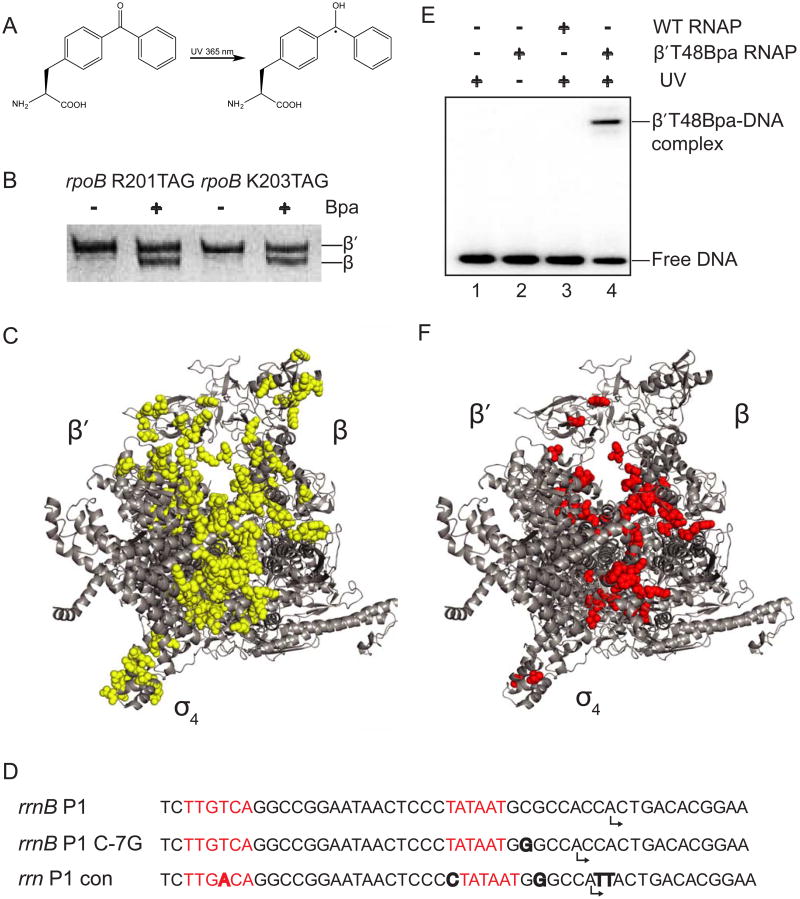
Figure 1. Identification of Bpa positions that crosslink to DNA.
(A) Structure of p-benzoyl-L-phenylalanine (Bpa). (B) SDS-PAGE of protein lysates from cells with TAG mutations at codons 201 or 203 of rpoB. Full-length β was only observed when core RNAP was overexpressed in cells grown in medium containing Bpa. (C) The 152 positions where Bpa substitutions were constructed in RNAP are shown as yellow spheres on a structure of E. coli RNAP holoenzymes (Zuo et al., 2013; PDB 4JKR; see also Table S1). Note that the wild-type amino acids are illustrated, not Bpa. (D) Sequences of rrnB P1 and the two promoters utilized here because they form more stable complexes with RNAP. The -10 and -35 hexamers are in red, differences from rrnB P1 are in bold, and predicted start sites are indicated by bent arrows. (E) Representative SDS-PAGE gel showing a 32P-labeled DNA fragment containing the rrnB C-7G promoter crosslinked to RNAP. Lane 1: no RNAP+UV. Lane 2: β′T48Bpa-RNAP no UV. Lane 3: WT RNAP +UV. Lane 4: β′T48Bpa-RNAP + UV. (F) 50 positions in E. coli RNAP where Bpa crosslinked to DNA are shown as red spheres. Related to Table S1.