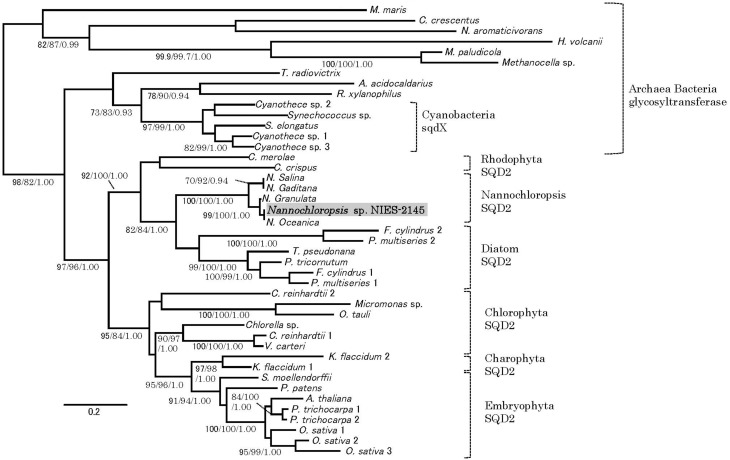
Figure 4.
Phylogenetic tree analysis of SQD2. A phylogeny incorporating eukaryotic SQD2, cyanobacterial sqdX and the glycosyl transferase proteins of other organisms was constructed using Bayesian inference, maximum likelihood and neighbor-joining algorithms. It is based on a protein comparison. The topologies and branch lengths were calculated using maximum likelihood. Bootstrap values (maximum likelihood and neighbor-joining) higher than 70 and Bayesian posterior probabilities (Bayesian inference) higher than 0.9 are indicated under each branch (maximum likelihood/neighbor-joining/Bayesian inference). The scale bar represents 0.2 amino acid substitutions per site. Sequence accession numbers or sequence resources for the phylogenetic tree are as follows: Archaea glycosyl transferase: Methanocella paludicola (YP_003355097), Methanocella sp. (YP_685710) and Haloferax volcanii (YP_003535220); Alpha-proteobacteria glycosyl transferase: Caulobacter crescentus (YP_002516166), Novosphingobium aromaticivorans (WP_011446186), and Maricaulis maris (WP_011642337); Cyanobacteria sqdX: Cyanothece sp. 1 (WP_015784192), Cyanothece sp. 2 (WP_012629991), Synechococcus elongatus (WP_011243256), Synechococcus sp. (WP_011429929), and Cyanothece sp. 3 (WP_015956855); Other bacterial glycosyl transferases: Truepera radiovictrix (WP_013178429), Rubrobacter xylanophilus (WP_011564279), and Alicyclobacillus acidocaldarius (WP_012810695); Rhodophyta SQD2: Chondrus crispus (XP_005715110), and Cyanidioschyzon merolae (XP_005538341); Stramenopile SQD2: Nannochloropsis sp. NIES-2145 (DDBJ Accession LC061442), Nannochloropsis oceanica IMET1 (BioProject: PRJNA202418, scaffold00247.g6721), Nannochloropsis granulata CCMP529 (BioProject: PRJNA65111, evm.model.NODE_3998_length_11613_cov_18.861534.4), Nannochloropsis salina CCMP537 (BioProject: PRJNA62503, evm.model.NODE_9973_length_179428_cov_24.782236.3), Nannochloropsis gaditana (EWM27152), Phaeodactylum tricornutum (JGI Protein ID: 50356), Fragilariopsis cylindrus 1 (JGI Protein ID: 207999), Fragilariopsis cylindrus 2 (JGI Protein ID: 158007), Pseudo-nitzschia multiseries 1 (JGI Protein ID: 252457), Pseudo-nitzschia multiseries 2 (JGI Protein ID: 145536), Thalassiosira pseudonana (JGI Protein ID: 38775), [Chlorophyta] Ostreococcus tauri (JGI Protein ID: 3203), Micromonas sp. (JGI Protein ID: 58169), Chlorella sp. (JGI Protein ID: 33086), Volvox carteri (Phytozome Transcript Name: Vocar20009459m), Chlamydomonas reinhardtii 1 (Phytozome Transcript Name: Cre01.g038550.t1.3), and Chlamydomonas reinhardtii 2 (Phytozome Transcript Name: Cre16.g689150.t1.2); Charophyta: Klebsormidium flaccidum 1 (BioProject: PRJDB718, kfl00392_0070) and Klebsormidium flaccidum 2 (BioProject: PRJDB718, kfl00041_0140), and Embryophyta (Phytozome Transcript Name): Arabidopsis thaliana (AT5G01220.1), Populus trichocarpa 1 (Potri.006G097600.1), Populus trichocarpa 2 (Potri.016G112600.1), Oryza sativa 1 (Os07g01030.1), Oryza sativa 2 (Os01g04920.1), Oryza sativa 3 (Os03g15840.1), Selaginella moellendorffii (170091), and Physcomitrella patens (Pp1s24_194V6.1).