Figure 3.
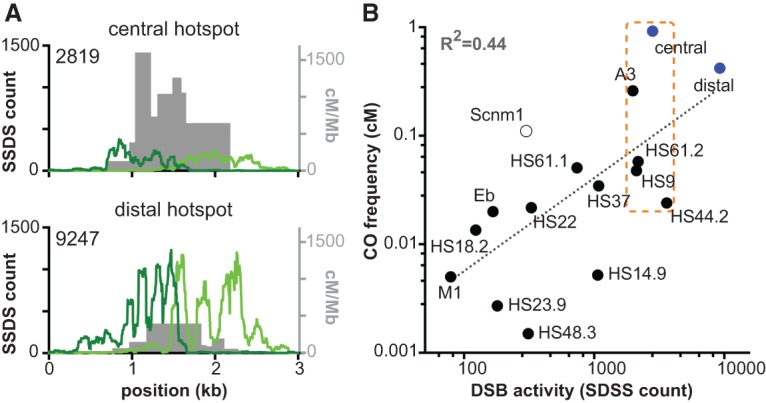
Estimates of relative DSB activity are an unreliable predictor of crossover frequency. (A) Comparison of SSDS reads with crossover distributions at the central and distal hot spots. Crossover breakpoint maps from spermatocytes (pooled from both orientations of allele-specific PCR) are shown in gray. Forward strand SSDS reads are shown in dark green, and reverse strand reads are in light green; total read counts (a measure of DSB activity) are indicated (data from Brick et al. 2012). The midpoint between accumulations of the forward and reverse strand reads marks the hot spot center. Note that A/J and B6 share the same Prdm9 allele, and the symmetry of crossover maps (Figs. 1C, 2A) indicates that recombination initiation occurs at comparable frequencies on both haplotypes. Thus, the SSDS and crossover maps are directly comparable even though they were generated from animals of different strain backgrounds (pure B6 for SSDS vs. A/J × B6 F1 hybrids for crossovers). Note that the Y-axis scales are the same for both hot spots. (B) Comparison of SSDS read counts and crossover frequencies at published mouse hot spots. SSDS data are from Brick et al. (2012). Filled blue circles are the hot spots from this study, filled black circles denote published hot spots assayed by allele-specific PCR of sperm DNA, and the open circle denotes a published hot spot assayed for crossing over by pedigree analysis (see Supplemental Table S5 for details). The dotted line is a least squares regression line fitted to the log transformed data. The orange box marks a group of hot spots that are within a factor of two of each other for SSDS signal but cover a much wider range in crossover activity. Note that different studies used different methods to correct recombination assays for amplifications efficiencies, so measured crossover frequencies may be underestimated to different degrees for specific hot spots (see the Materials and Methods for further details).
