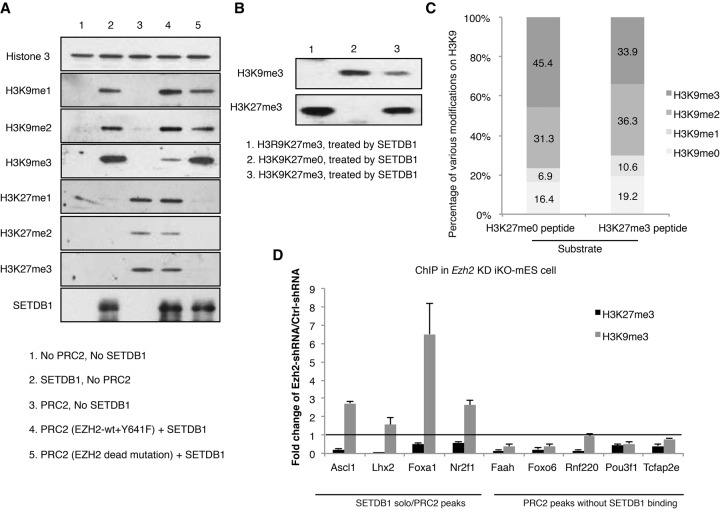
Figure 4.
H3K27 methylation inhibits SETDB1-mediated H3K9 methylation. (A) In vitro methylation assay was performed using recombinant nucleosomes. The recombinant nucleosome was first reacted with EZH2 (1:1 mixture of wild-type and the Y641F mutant EZH2 protein together with EED, RBBP4, AEBP2, and SUZ12) to induce H3K27 methylation. Recombinant human SETDB1 protein was then added to the reaction for 4 h at room temperature. The reaction was measured by Western blot analysis using antibodies against various H3K9 methyl marks, as well as H3K27 methyl marks. SETDB1 Western blot analysis confirmed the equal amount of the SETDB1 protein in the reaction. (B) Chemically modified H3 (1-84aa) peptides were used as the substrates in the in vitro methylation assay with SETDB1. Peptides with H3K27me3 modification (lane 3) showed reduced H3K9 trimethylation by SETDB1 in vitro compared to H3K27me0 peptides (lane 2). A peptide with K9 mutated to R was used as the control (lane 1). (C) The end products of the peptide-based, in vitro methylation assay were quantified by LC/MS analysis. The percentage of the various modifications was shown in the stack bar graphs. The left and right plots correspond to the product from the peptide substrate with (lane 3 in B) or without H3K27me3 (lane 2 in B) modification, respectively. Note that the H3K9me3 production is dramatically reduced when the H3K27me3 peptide was used as the substrate. (D) Setdb1 iKO cells were infected with a lentiviral shRNA against Ezh2 or control scramble shRNA. The chromatin was immunoprecipitated with antibodies against H3K27me3 or H3K9me3 histone marks. The data are normalized to the control shRNA samples. Four representative SETDB1 solo/PRC2 loci were analyzed by qPCR using the immunoprecipitated DNA. The selected SETDB1 solo/PRC2 peak loci were marked by their nearby gene symbols (Ascl1, Lhx2, Foxa1, and Nr2f1). Five PRC2 peaks without SETDB1 binding were used as the control. The selected PRC2 peak loci without SETDB1 binding were marked by their nearby gene symbols (Faah, Pou3f1, Tcfap2e, Rnf220, and Foxo6).