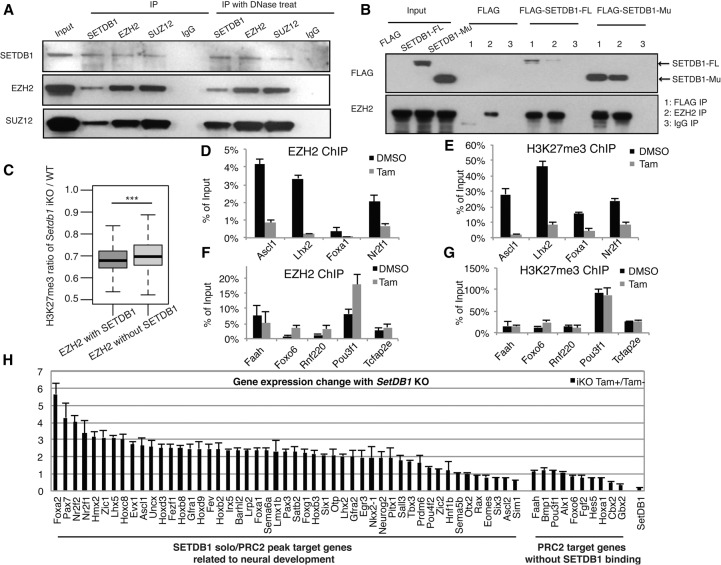
Figure 5.
SETDB1 modulates H3K27 methylation and EZH2 binding. (A) Coimmunoprecipitation (co-IP) was performed to validate SETDB1 and PRC2 correlation. Nuclear proteins of SETDB1 iKO-ES cells were extracted for IP with SETDB1, EZH2, and SUZ12 antibodies. IgG was used as a negative control. Western blot analysis was performed with SETDB1, EZH2, and SUZ12 antibodies. Cell cytoplasmic and nuclear fractions were used as the input controls. (Left) co-IP results without DNase treatment; (right) those with DNase treatment prior to the IP procedure. (B) 3xFLAG-tagged, full-length wild-type or SET domain-deleted SETDB1 was transfected into iKO mouse ES cells. Nuclear proteins were extracted for IP using the antibodies for SETDB1 or EZH2. IgG was used as a negative control. (C) H3K27me3 level variation at all ±10-kb regions of EZH2 peak loci before and after Setdb1 iKO (biological replicate pair 1). Only EZH2 peaks with enriched H3K27me3 signal before Tam treatment were included in this analysis. Those EZH2 peaks were divided into two categories based on the overlap status with SETDB1 binding: with SETDB1 binding (431 peaks) and without SETDB1 binding (13,138 peaks). Two-sided Welch's t-test was performed for the change of H3K27me3 between the two groups; (***) P < 0.001. Supplemental Figure S12 showed the results using biological replicate pair 2. (D) ChIP-qPCR of EZH2 in selected SETDB1 solo/PRC2 peak loci upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-EZH2 antibody. The selected SETDB1 solo/PRC2 peak loci were marked by their nearby gene symbols (Ascl1, Lhx2, Foxa1, and Nr2f1). (E) ChIP-qPCR of H3K27me3 in selected SETDB1 solo/PRC2 peak loci upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-H3K27me3 antibody. (F) ChIP-qPCR of EZH2 in selected PRC2 peak loci without SETDB1 binding upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-EZH2 antibody. The selected PRC2 peak loci without SETDB1 binding were marked by their nearby gene symbols (Faah, Foxo6, Rnf220, Pou3f1, and Tcfap2e). No reduction of EZH2 binding was observed. (G) ChIP-qPCR of H3K27me3 in selected PRC2 peak loci without SETDB1 binding upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-H3K27me3 antibody. No reduction of H3K27me3 enrichment was observed. (H) Setdb1 iKO ES cells were treated with or without Tam for 3 d. Setdb1 expression level was detected by qPCR for knockout efficiency confirmation. The expression level of the 49 SETDB1 solo/PRC2 peak target neural development–related genes was examined by RT-qPCR analysis. Ten PRC2 target genes without SETDB1 binding were used as the negative control. Please note that while there is significant reactivation of the SETDB1 solo/PRC2 peak target genes related to neural development (P-value <0.05, paired t-test) as a group, there is no consistent change of the PRC2 target genes without SETDB1 binding.