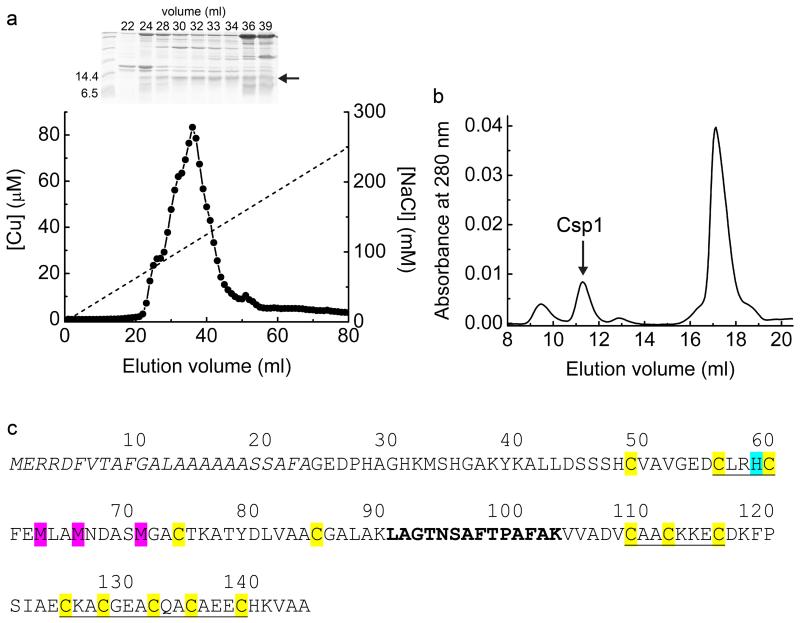
Extended Data Figure 1 . Purification of proteins from M. trichosporium OB3b and the amino-acid sequence of Csp1.
a, The copper content of anion-exchange fractions (NaCl gradient shown as a dashed line) and the SDS-PAGE analysis of selected fractions (1 mL) from the purification of soluble extract from M. trichosporium OB3b cells. The band just below the 14.4 kDa marker, indicated with an arrow, is present. Fraction 32 was judged to have the lowest level of contaminating proteins and was further purified by gel-filtration chromatography on a Superdex 75 column (b). Csp1 is present in the peak that elutes at ~ 11 mL and contains considerable copper (see Fig. 1c). c, The amino-acid sequence of Csp1 showing the predicted Tat leader peptide (the first 24 residues of the pre-protein) in italics. The 13 Cys residues are highlighted in yellow and His36 (cyan), Met40, Met43 and Met48 (magenta) are also indicated (the numbering of these residues refers to the mature protein). The CXXXC and CXXC motifs are underlined. The region in bold corresponds to the single tryptic fragment identified on two separate occasions in MS analysis, representing 11% sequence coverage of the mature protein (Mascot search of peptide mass fingerprint, expect value = 1.9 × 10−5). The sequence of this fragment was confirmed by liquid chromatography/MS/MS (data not shown). This is the only tryptic peptide from the mature protein that would be anticipated to be readily detected by MS (due to either small mass or presence of Cys residues in all other theoretical tryptic fragments) and is unique to this protein amongst all proteobacterial protein sequences in the NCBInr database.