Figure 11.
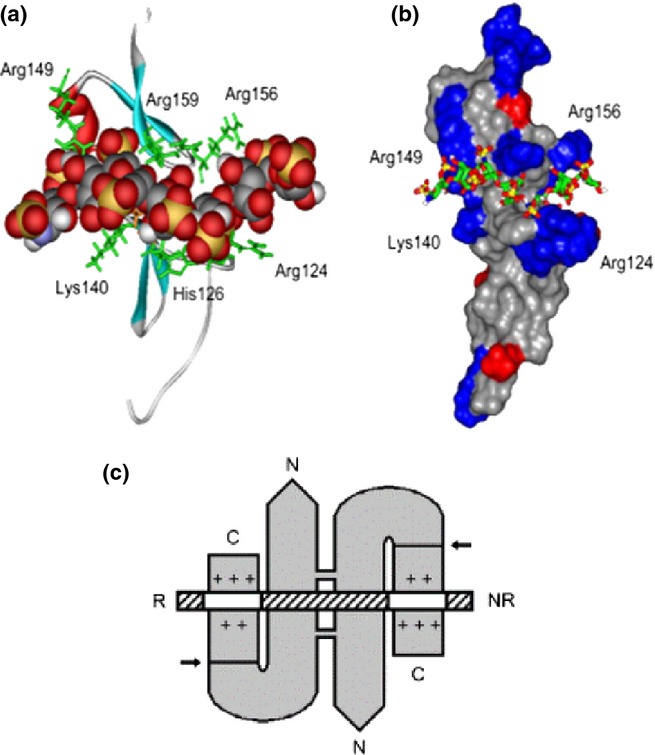
Heparin-/heparan sulphate (HS)-binding sites in the VEGF165 heparin-binding domain (HBD) and in the VEGF165 dimer. (a) Ribbon diagram of the VEGF165 HBD (residues 111–165; Protein Data Bank code 2VGH) with a docked heparin dp7 [space-filling representation: carbon (grey), oxygen (red), sulphur (yellow), nitrogen (blue) and hydrogen (white)]. Basic residues lining the shallow binding groove are shown in a stick representation (green). (b) The same complex with 2VGH depicted as a protein surface. Arg and Lys residues are shown in blue, and Glu and Asp residues are shown in red. The heparin dp7 saccharide is a stick representation. The atomic coloration is as in (a), except that carbons are shown in green. (c) A K5 lyase-resistant HS fragment (white and hatched boxes) is bound to the HBDs of VEGF165 homodimer (grey). The N- and C-termini of VEGF165 and the reducing (R) and nonreducing (NR) ends of the HS chain are indicated. The VEGF165 subunits are held by disulfide bonds in an antiparallel ‘side-by-side’ orientation. Arrows indicate plasmin cleavage at the sites that release the HBDs. Basic residues in each heparin-binding cleft are shown (+). The two clefts are occupied by separate S domains (white boxes) in the same HS chain. The S domains are at least dp6 in length and are 6-O-sulphated. This research was originally published in Journal of Biological Chemistry. Robinson et al. 2006 © the American Society for Biochemistry and Molecular Biology.” VEGF, vascular endothelial growth factor.
