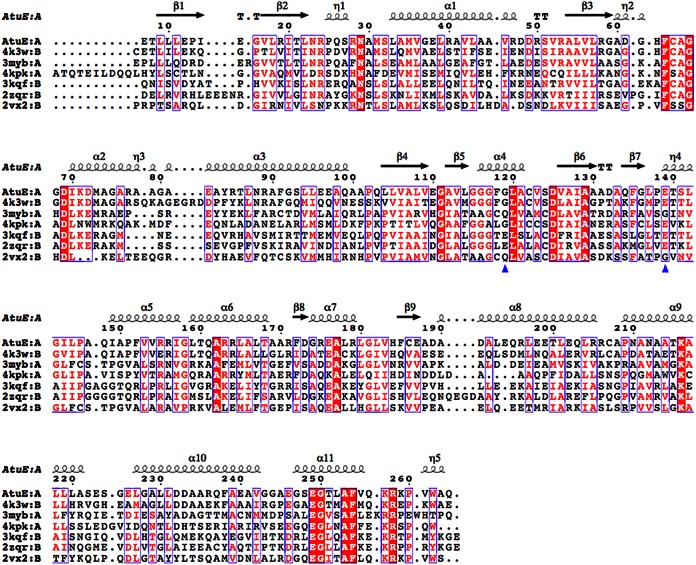
FIG 3.
Structure-based alignment of AtuE homologs. Only the top six structures are listed in descending order of the Q score (from 0.88 to 0.71). The residues corresponding to rat ECH catalytic glutamates are shown with a blue triangle. Details of the structures and their structure-based sequence identities to AtuE are as follows: the structure with PDB accession number 4K3W (Marinobacter aquaeolei enoyl-CoA hydratase), 56% sequence identity; the structure with PDB accession number 3MYB (Mycobacterium smegmatis enoyl-CoA hydratase), 29% sequence identity (Seattle Structural Genomics Center for Infectious Disease); the structure with PDB accession number 4KPK (Shewanella pealeana ATCC 700345), 32% sequence identity (New York Structural Genomics Research Consortium); the structure with PDB accession number 3KQF (Bacillus anthracis enoyl-CoA hydratase), 31% sequence identity (G. Minasov, A. Halavaty, Z. Wawrzak, T. Skarina, O. Onopriyenko, L. Papazisi, A. Savchenko, and W. F. Anderson, structure to be published); the structure with PDB accession number 2ZQR (human AU RNA binding protein/enoyl-coenzyme A hydratase), 27% sequence identity (35); and the structure with PDB accession number 2VX2 (human enoyl-CoA hydratase domain-containing protein 3), 21% sequence identity (W. W. Yue, K. Guo, G. Kochan, E. Pilka, J. W. Murray, E. Salah, R. Cocking, Z. Sun, A. K. Roos, A. C. W. Pike, P. Filippakopoulos, C. Arrowsmith, M. Wikstrom, A. Edwards, C. Bountra, and U. Oppermann, structure to be published). The homologous hydratases have a trimer composition according to the PDB. The figure was created with the ESPript program (version 3.0) (36). The structure-based alignment was carried out by PDBeFold. Conserved residues are indicated by white letters on a red background (strictly conserved) or red letters on a white background (global similarity score, >0.7) and framed in blue boxes.