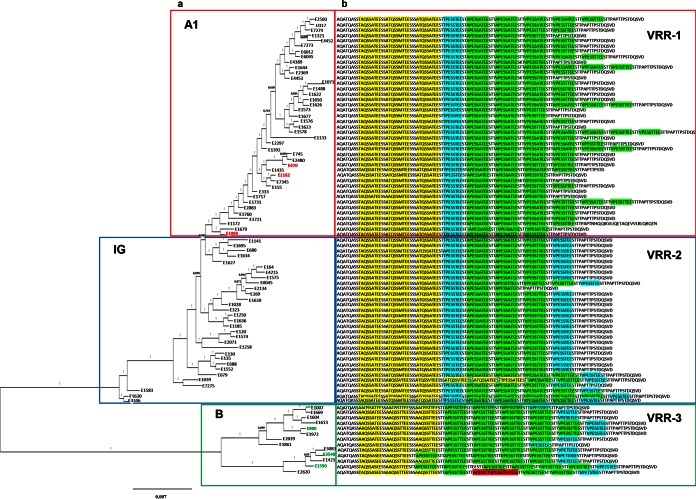
FIG 1.
Phylogenetic reconstruction of 85 E. faecium strains based on core genome variation. (a) Phylogenetic tree (FastTree) built from an alignment of 1,186 core genes in 85 E. faecium genomes. Clades A1 and B and the intermediate group (IG) are indicated by red, green, and blue boundaries, respectively. Selected strains for further phenotypic tests are highlighted in red in clade A1 (E470, E1162, and E1904) and in green in clade B (E980, E3548, and E1590). (b) SagA variable repeat regions (VRRs) are indicated after each strain designation. The VRRs are divided into three types: VRR-1 (boxed in red), VRR-2 (boxed in blue), and VRR-3 (boxed in green). The sequences highlighted in yellow, blue, green, and red indicate the different repeat variants.