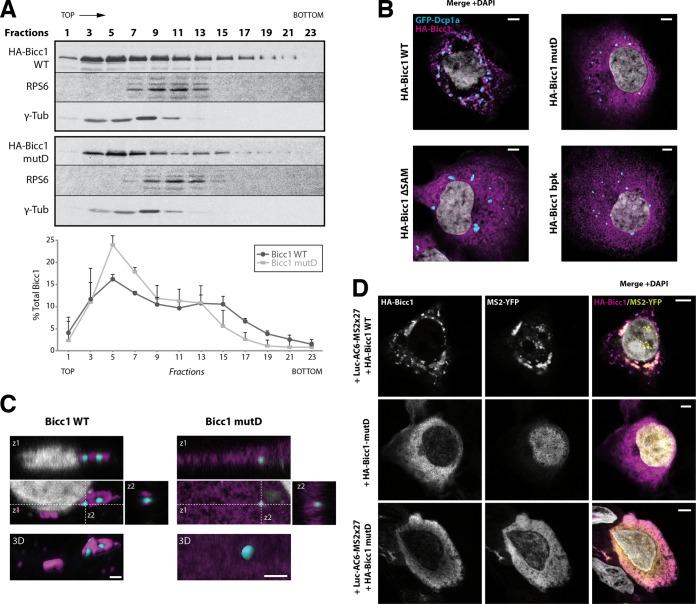
FIG 5.
SAM polymerization is required for Bicc1 clustering. (A) Density fractionation of WT and polymerization mutant Bicc1 on a sucrose gradient. HEK293T cell extracts containing HA-tagged Bicc1 were fractionated on a continuous 15 to 60% sucrose gradient and analyzed by anti-Bicc1 Western blotting. The migration direction from the top to the bottom of the tube is indicated. RPS6 and γ-tubulin (γ-Tub) were used as internal controls. The graph below the gels shows the percentage of Bicc1 compared to the total Bicc1 signal for each fraction. Results represent mean values from 3 independent experiments, and error bars show SEMs. (B) Bicc1 polymer mutants fail to accumulate in cytoplasmic foci. The results of indirect immunofluorescence staining of the HA-Bicc1 WT, mutD, the ΔSAM mutant, or the bpk mutant and the P-body marker GFP-Dcp1a overexpressed in COS-1 cells are shown. Bars, 5 μm. (C) Comparative 3D rendering of the HA-Bicc1 WT and mutD by Imaris software. From the original image (center), z-stacks in two directions (z1 and z2, top and right, respectively) and 3D reconstruction (bottom) are given. Bars, 2 μm. (D) Localization by indirect immunofluorescence staining of the Luc-AC6-MS2×27 reporter mRNA and HA-Bicc1 in COS-1 cells and comparison with that of HA-Bicc1 mutD. Bars, 5 μm.