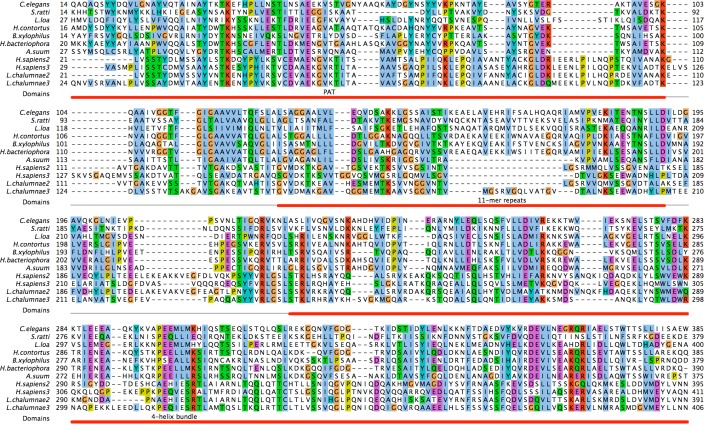
Figure 1. Identification of C. elegans protein W01A8.1a as a close homologue of vertebrate perilipin.
C. elegans protein W01A8.1a is compared with nematode homologues of pairwise sequence identity lower then 70% and with Plin2 and 3 from two diverse vertebrates. The three perilipin specific domains (indicated in red) were identified through homology with human Plin3. The six 11-mer repeats in W01A8.1a (positions 126–136, 137–147, 148–158, 159–169, 170–180 and 181–191) were established with HHrepID algorithm (Biegert & Soding, 2008). The N-terminal PAT domain is thought to interact with HSL. The central domain consisting of imperfect 11-mer repeats forming amphipathic helices is responsible for the main affinity to LDs and the C-terminal domain containing an apolipoprotein-like 4-helix bundle probably plays an additional role in the affinity to LDs and is known to interact with ABHD5 in mammalian Plin1 and 3 (Brasaemle, 2007). Alignment was done using T-coffee alignment of all available nematode sequences aligned with vertebrate Plin2 and 3 sequences in three iterations using ProfileAlign routine in MyHits suite (myhits.isb-sib.ch). Selected sequences from top to bottom: (Species, database identifier): Caenorhabditis elegans, Q23095; Strongyloides ratti, CACX01001972.1; Loa loa, E1G5Y0 and ADBU02007219.1; Haemonchus contortus, CDJ80228.1; Bursaphelenchus xylophilus, CADV01008520.1; Heterorhabditis bacteriophora, ES742365.1 and ACKM01001830.1; Ascaris suum, U1NU60; Homo sapiens 2, PLIN2_HUMAN; Homo sapiens 3, PLIN3_HUMAN; Latimeria chalumnae 2, H3AYC0; Latimeria chalumnae 3, GAAA01019375.1. Nucleotide sequences were translated with Wise2 program (Birney, Clamp & Durbin, 2004). Amino acid types are colored according to the Clustal scheme (jalview.org/help/html/colourSchemes/clustal.html).