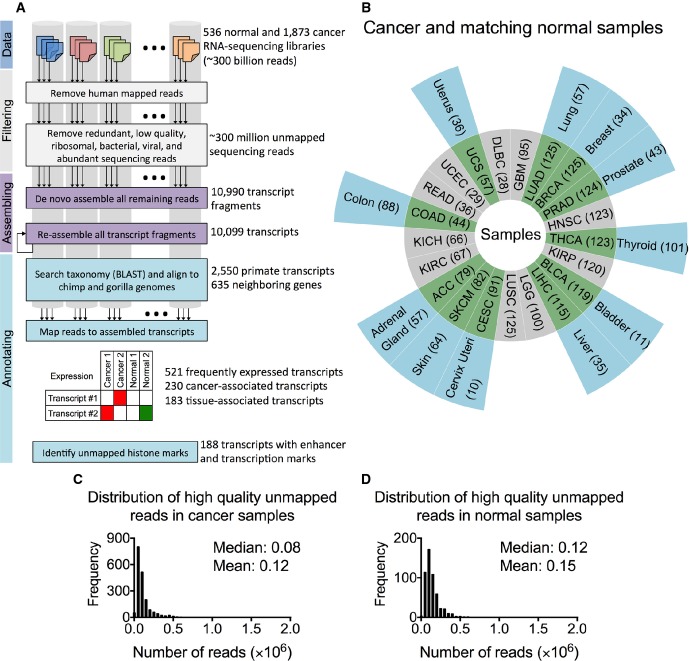
Figure 1. Characterizing unmapped sequences.
- A Data processing strategy for identifying missed transcripts. Sequencing reads from cancer and normal samples were mapped to the human genome and transcriptome. Abundant reads (e.g., polyA, polyC, ribosomal RNAs, phage) and low-quality reads were discarded, and reads that mapped to known viral and bacterial sequences were removed. The remaining unmapped reads were pooled and de novo assembled to obtain previously missed transcripts. The newly assembled transcripts were annotated by their over- or under-representation in each cancer and the presence of histone marks in their genomic loci. For the illustrated 2 transcripts, one was expressed only in cancer and one in both cancer and normal tissues.
- B Cancer types (inner donut) and matching normal tissue (outer donut). The numbers of samples are in parentheses. The abbreviations for the different cancer types are in Table EV1A.
- C, D Distribution of high-quality unmapped sequencing reads across all cancer (C) and normal (D) samples after screening as described in (A).