Abstract
Background/Aims
To screen for serum protein/peptide biomarkers of hepatitis B virus (HBV)-associated chronic hepatic lesions in an attempt to profile the progression of HBV-associated chronic hepatic lesions using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS) techniques.
Methods
Using SELDI-TOF MS, serum protein/peptide profiles on the CM10 ProteinChip arrays were obtained from a training group including 26 HBV-associated hepatocellular carcinoma patients with liver cirrhosis (LC), 30 HBV-associated LC patients, 85 patients at different stages of liver fibrosis, and 30 asymptomatic HBV carriers. The most valuable SELDI peak for predicting the progression to LC in HBV-infected patients was identified.
Results
A SELDI peak of M/Z 5805 with value for predicting LC in HBV-infected patients was found and was identified as a peptide of the C-terminal fraction of the fibrinogen α-chain precursor, isoform 1.
Conclusions
The peptide of the C-terminal fraction of the fibrinogen α-chain precursor, isoform 1 with M/Z 5805, may be a serological biomarker for progression to LC in HBV-infected patients.
Keywords: Chronic hepatic lesions, Proteomics, Diagnosis, Surface-enhanced laser desorption/ionization, Biological markers
INTRODUCTION
Chronic hepatitis B virus (HBV) infection is the most common cause of liver disease, leading to long-term liver damage including fibrosis, cirrhosis, and hepatocellular carcinoma (HCC).1,2 Chronic hepatitis B infects approximately 400 million people worldwide and causes 1 million deaths of liver disease per year.1 Clinically, people with chronic HBV hepatitis are at high risk of liver damages, with approximately 15% to 40% of infected population experiencing chronic liver damages, liver failure or HCC if not treated properly at the early stage.3
Liver biopsy is now recommended as the gold standard method of staging chronic hepatic diseases with HBV infection despite of its disadvantages such as poor compliance to invasive method, sampling error, limited usefulness for dynamic surveillance and follow-up. So there is still a need for the development of noninvasive techniques to monitor the clinic stage of the liver damages in view of the above-mentioned limitations and reluctance.
Currently, the development of innovative technologies in genomic and proteomic analysis of HBV-related chronic hepatic diseases might bring great hopes that the techniques not only contribute to our understanding of the diseases, but also help to develop diagnostic markers that could assist the clinicians in their management.4 At present, it has become relatively easy to detect the protein profiling in the crude biological samples with surface enhanced laser desorption/ionization-time of flight mass spectrometry (SELDI-TOF MS).5 The most common use of the SELDI-TOF MS today is in biomarker researches by comparing serum proteins between healthy and diseased individuals.6,7 This is a high-throughput technical platform which can detect multiple protein changes simultaneously with high sensitivity and specificity.8,9
In the present study, by comparing analysis among normal volunteers and patients suffering from different chronic hepatic diseases associated with HBV infection using Ciphergen SELDI Software 3.1.1 (Ciphergen Biosystems, Fremont, CA, USA) with Biomarker Wizard, some serum proteins were identified as new candidate biomarkers for the prediction of progression patterns of these hepatic diseases. At the same time, the most valuable M/Z 5805 protein was validated as the marker for monitoring hepatic fibrosis and cirrhosis.
MATERIALS AND METHODS
1. Patients
Four groups of consecutive subjects were enrolled in this research. (1) HCC group: This group included 26 HBV-associated HCC patients based on liver cirrhosis (LC). Eligible criteria included pathologically proved HBV associated LC (Ishak=6) and excluded other cofactor. (2) HBV associated LC group: This group included 30 HBV-associated LC (Ishak=6). Eligible criteria included pathologically proved HBV associated LC and excluded other cofactor. (3) Chronic HBV infection group: This group included 30 patients with liver fibrosis stage I (Ishak score=1), 27 patients with liver fibrosis stage II (Ishak score=2, 3), 14 patients with liver fibrosis stage III (Ishak score=4) and 14 patients with liver fibrosis stage IV(Ishak score=5). Eligible criteria included pathologically proved and excluded other cofactor. (4) Control group: The third group included 30 HBV asymptomatic carriers by excluded other virus infection or alcohol intake. The demographic information of all subjects is shown in Table 1. The patients from HCC group, LC group, chronic liver disease group were all proved by pathological diagnosis.
Table 1.
Demographic Information for All Groups
| Group | Age, yr | Gender, M/F | |
|---|---|---|---|
|
| |||
| Range | Mean±SD | ||
| Control | 22–56 | 36.4±13.19 | 27/5 |
| LF I | 22–53 | 34±9.31 | 28/2 |
| LF II | 23–60 | 38.3±9.48 | 21/6 |
| LF III | 27–53 | 36.2±8.28 | 8/5 |
| LF IV | 27–54 | 38.4±8.18 | 13/1 |
| LC | 34–66 | 48.5±9.67 | 23/7 |
| HCC | 35–70 | 55.7±11.9 | 23/2 |
M, male; F, female; LF, liver fibrosis; LF I, hepatitis B virus (HBV)-associated liver fibrosis stage I; LF II, HBV-associated liver fibrosis stage II; LF III, HBV-associated liver fibrosis stage III; LF IV, HBV-associated liver fibrosis stage IV; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
2. Samples
All blood samples of study population were collected from Changzheng Hospital (Shanghai, China) and Chinese PLA General Hospital (Beijing, China). The experimental protocol was approved by the Health Human Research Ethics Committee of Changzheng Hospital and Chinese PLA General Hospital. All the blood samples were collected in the fasting state in the early morning. No patient received blood transfusion within 2 months before collection of the sera. Blood samples were stably stored at 4°C for 1 hour, and then centrifuged at 1,400 g for 15 minutes at 4°C to separate the sera, and harvested sera were then stored at −80°C until use. Inform consent was obtained from all the patients for blood sampling.
3. Serum proteomic profiling
All serum samples were processed and assayed blinded by investigator. The SELDI ProteinChip analysis was performed with CM10 ProteinChip arrays at a pH of 3. Six microliters of the serum sample were denatured by adding 12 μL of U9 solution (9 mol/L urea, 2% 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate, 50 mmol/L Tris-HCl, pH 9), and diluted with 222 μL of binding buffer (150 mmol/L sodium acetate, 0.1% Triton X-100, pH 3.0) for CM10 ProteinChip array experiments to result in a final dilution of 20-fold. Briefly, the array spots were preactivated with binding buffer (150 mmol/L sodium acetate, pH 3.0; Sigma, Santa Clara, CA, USA) for 5 minutes at room temperature in duplicate, followed by two washes with deionized water. Then 200 μL of the diluted sample were applied to the pre-equilibrated ProteinChip array and incubated with shaking at room temperature for 90 minutes. After the incubation, each array was washed twice with the binding buffer and rinsed twice with deionized water. After air drying, sinapinic acid matrix in 50% acetonitrile and 0.5% trifluoroacetic acid was added to each array.
The arrays data were analyzed with the Ciphergen Protein-Chip Reader (model PBS II). The mass spectra of proteins were generated by using an average of 160 laser shots. The detection sizes ranged between 2 kd and 20 kd and a 30-shot average per sample. Sensitivity of the detector was set at 9. The mass-to-charge ratio (m/z) of each protein captured on the array surface was determined upon externally calibrated standards (Ciphergen Biosystems) with the 0.1% mass accuracy of the spectrometer according to the manufacturer.
4. Peak detection
Ciphergen ProteinChip software was used for automatic experimental data collection, and Biomarker Wizard 3.1 software was used to analyze the variability of the protein peaks. The settings used for the work were as follows: for peak detection, the signal-to-noise ratio was 5, and the minimum peak threshold was 10%.
5. Peak identification
Serum peptides were enriched and purified from the mixture of the sera of 10 HCC patients with high 5805 peak by using the Weak Cation Exchanger (WCX) Clinprot magnetic beads (Bruker Daltonics, Billerica, MA, USA). Samples (20 μL, 2 μL each patient) were purified through three steps—binding, washing, and elution—according to the manufacturer’s suggested protocol, in which the binding incubation time took 1 minute.
The solutions that were eluted from the magnetic beads were purified using Michrome Protein MicroTrap (Michrom Bio-Resources, Auburn, CA, USA) to remove the impurities, using the procedure described by the manufacturer. All the peptide solutions above were dried thoroughly using a vacuum centrifuge and then resuspended with 5% ACN in 0.1% formic acid, separated by nanoLC and analyzed by online electrospray tandem mass spectrometry. The experiments were performed on a Nano Aquity UPLC system (Waters Corp., Milford, MA, USA) connected to an LTQ Orbitrap XL mass spectrometer (Thermo Electron Corp., Bremen, Germany) equipped with an online nanoelectrospray ion source (Michrom Bioresources). The separation of the peptides was performed in a Symmetry®C18, 5 μm, 180 μm id×2 cm trap-column and a BEH300 C18, 3.5 μm, 75 μm id×15 cm reverse phase column (Waters Corp.). The peptide mixtures were injected onto the trap-column with a flow of 15 μL/min for 3 minutes and subsequently eluted with a three-step linear gradient. Starting from 5% B to 45% B in 40 minutes (A: water with 0.1% formic acid; B: ACN with 0.1% formic acid), increased to 80% B in 1 minute, and then hold on 80% B for 4 minutes. The column was re-equilibrated at initial conditions for 15 minutes. The column flow rate was maintained at 400 nL/min and column temperature was maintained at 35°C. The electrospray voltage of 1.8 kV versus the inlet of the mass spectrometer was used.
LTQ Orbitrap XL mass spectrometer was operated in the data-dependent mode to switch automatically between MS and MS/MS acquisition. Survey full-scan MS spectra with two microscans (m/z 400–2000) were acquired in the Obitrap with a mass resolution of 100,000 at m/z 400, followed by 10 sequential LTQ-MS/MS scans. Dynamic exclusion was used with two repeat counts, 10-s repeat duration, and 30-s exclusion duration. For MS/MS, precursor ions were activated using 25% normalized collision energy at the default activation q of 0.25. All MS/MS spectrums were identified by using International Protein Index (IPI human v3.45 fasta with 71983 entries).
6. Data analysis
The data were analyzed with ProteinChip Software version 3.1.1 (Ciphergen Biosystems). For each comparison, the raw intensity data were normalized by using the total ion current of all profiles. The peak intensities were normalized to the total ion current of m/z between 2,000 and 20,000 Da for the low-molecular weight range.
The protein profiling of all chronic hepatic disease with HBV infection group were compared each to find significantly different protein peaks for progression chronic hepatic disease with HBV infection using the Wilcoxon rank test. Mean peak intensity of each protein was calculated and compared. The receiver operator characteristic curves were made using fisher linear discrimination analysis by S-PLUS 6.2 Professional Software. The classification performance (specificity and sensitivity) of between each two groups were assessed by the area under the curve of the receiver operator characteristic curves.
RESULTS
1. Study population
The demographic information of the patients was showed in Table 1.
2. Screening and analysis of diverse serum proteins related to chronic hepatic diseases with HBV infection
Through Biomarker Wizard 3.1 software, 294 protein peaks were detected among the four groups of by using principle that the 20% cases have the peak signal to noise ratio higher than five (S/N>5). There are 18 proteins have obviously deviation in the compare among control group, liver fibrosis group (stage I–IV), LC group and HCC group (p<0.05), respectively. The average of the peak in different groups was showed in Table 2.
Table 2.
Peaks Deviated in Chronic Hepatic Diseases Associated with Hepatitis B Virus Infection
| M/Z | p | Intensity of control | Intensity of LF I | Intensity of LF II | Intensity of LF III | Intensity of LF IV | Intensity of LC | Intensity of HCC |
|---|---|---|---|---|---|---|---|---|
| 5805 | 4.22×10−6 | 1.11±0.53 | 1.15±0.55 | 1.60±0.87 | 1.81±0.93 | 2.05±1.15 | 9.17±1.30 | 11.63±1.87 |
| 13260 | 4.07×10−5 | 1.49±0.73 | 1.61±0.66 | 1.29±0.41 | 0.81±0.37 | 0.80±0.32 | 0.33±031 | 0.62±0.52 |
| 8913 | 6.82×10−5 | 2.96±1.32 | 5.99±1.35 | 4.91±1.69 | 5.87±1.50 | 5.37±1.80 | 4.18±1.37 | 3.58±1.23 |
| 8767 | 1.25×10−4 | 5.18±1.54 | 7.55±2.13 | 8.07±1.75 | 8.36±1.56 | 6.41±1.57 | 4.59±1.68 | 4.89±1.44 |
| 13755 | 4.12×10−4 | 2.62±1.35 | 3.11±1.72 | 3.49±1.18 | 3.04±1.62 | 1.80±1.05 | 1.15±0.89 | 1.28±1.00 |
| 11687 | 7.48×10−4 | 0.60±0.30 | 1.61±0.53 | 1.28±0.84 | 1.57±1.00 | 1.45±0.84 | 4.55±2.70 | 1.71±0.96 |
| 8689 | 9.65×10−4 | 37.96±9.71 | 49.69±11.37 | 50.56±10.28 | 54.35±15.81 | 45.91±19.00 | 38.03±16.53 | 38.33±12.05 |
| 12858 | 2.31×10−3 | 1.59±0.57 | 2.46±0.98 | 2.28±0.91 | 2.75±0.48 | 1.97±0.93 | 0.87±0.68 | 1.09±0.82 |
| 8563 | 4.03×10−3 | 20.43±11.52 | 25.57±9.29 | 27.39±6.49 | 26.26±5.60 | 23.51±5.17 | 12.84±6.29 | 17.69±2.16 |
| 13062 | 4.50×10−3 | 1.43±0.26 | 1.60±0.98 | 1.28±0.91 | 1.38±0.68 | 0.99±0.48 | 0.45±0.22 | 0.84±0.26 |
| 9190 | 4.82×10−3 | 17.64±4.53 | 5.60±1.88 | 13.49±8.49 | 0.21±0.32 | 3.36±1.32 | 5.45±4.35 | 16.85±3.50 |
| 4348 | 4.91×10−3 | 13.45±5.74 | 20.82±6.75 | 17.64±7.96 | 23.24±10.05 | 20.52±9.88 | 13.90±7.23 | 12.65±7.12 |
| 3318 | 1.02×10−2 | 45.95±11.62 | 50.59±11.00 | 46.44±12.55 | 49.93±14.93 | 59.54±12.99 | 32.99±17.60 | 31.75±14.81 |
| 6837 | 1.55×10−2 | 18.09±6.80 | 22.27±7.11 | 22.70±8.17 | 21.87±7.31 | 24.24±6.39 | 16.97±8.38 | 14.52±6.93 |
| 3421 | 2.60×10−2 | 8.77±4.42 | 9.93±3.52 | 8.01±3.87 | 8.67±4.09 | 9.57±4.66 | 5.23±4.16 | 5.08±3.02 |
| 5987 | 3.14×10−2 | 3.21±2.77 | 3.73±2.45 | 3.62±1.60 | 3.72±1.23 | 2.76±0.71 | 2.12±0.59 | 2.45±0.75 |
| 5338 | 3.33×10−2 | 5.82±1.25 | 2.91±1.61 | 3.21±103 | 2.86±1.93 | 2.84±1.43 | 12.15±4.47 | 13.73±5.12 |
| 3933 | 3.50×10−2 | 3.29±2.64 | 0.62±0.47 | 0.87±0.44 | 1.12±0.78 | 0.23±0.25 | 1.17±0.78 | 6.78±3.99 |
Data are presented as mean±SD.
LF, liver fibrosis; LF I, hepatitis B virus (HBV)-associated liver fibrosis stage I; LF II, HBV-associated liver fibrosis stage II; LF III, HBV-associated liver fibrosis stage III; LF IV, HBV-associated liver fibrosis stage IV; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
Among the 18 proteins, 5805 Da protein (p<0.0001) is the most significant peak. Our data indicated that 5805 Da protein is undetected in control group while expressed upward from control to liver fibrosis (stage I–IV), LC and HCC, with the big hoist in LC and HCC groups (p=0.003) (Table 3, Fig. 1). In this way, 5805 Da protein could be a potential biomarker for identifying LC from fibrosis stage IV and predict the progress of HBV-related liver diseases. The diagnostic sensitivity and specificity reach 73.3% and 85%, respectively when 5805 Da peak is applied as the biomarker for distinguishing LC (Ishak=6) from fibrosis stage IV (Ishak=5). The area under the receiver characteristic curve was 0.77 (Fig. 2). The cutoff value was Intensitym/z=5805 3.6.
Table 3.
The Mean Intensity of the 5805-Da Peak among All Groups
| Group | X̄m/z | Intensity |
|---|---|---|
| Control | 5805.89 | 1.11±0.53 |
| LF I | 5805.40 | 1.15±0.55 |
| LF II | 5805.37 | 1.60±0.87 |
| LF III | 5806.11 | 1.81±0.93 |
| LF IV | 5805.27 | 2.05±1.15 |
| HBV-associated LC | 5804.41 | 9.17±1.30 |
| HBV-associated HCC | 5805.49 | 11.63±1.87 |
LF, liver fibrosis; LF I, hepatitis B virus (HBV)-associated liver fibrosis stage I; LF II, HBV-associated liver fibrosis stage II; LF III, HBV-associated liver fibrosis stage III; LF IV, HBV-associated liver fibrosis stage IV; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
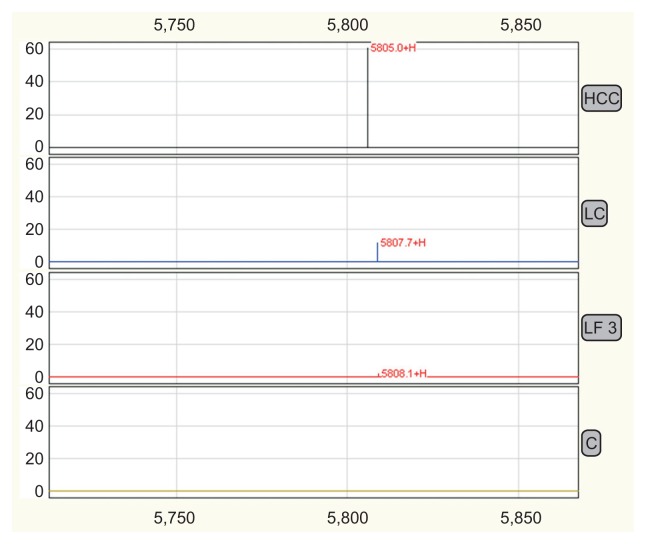
Fig. 1.
The 5805 peak profiling in the control, liver fibrosis (LF), liver cirrhosis (LC), and hepatocellular carcinoma (HCC) groups showed that the intensity of the 5805 peak was much higher in the LC and HCC groups. The 5805-Da protein is a potential biomarker for differentiating LC from stage IV fibrosis and predicting the progression of hepatitis B virus-related liver diseases.
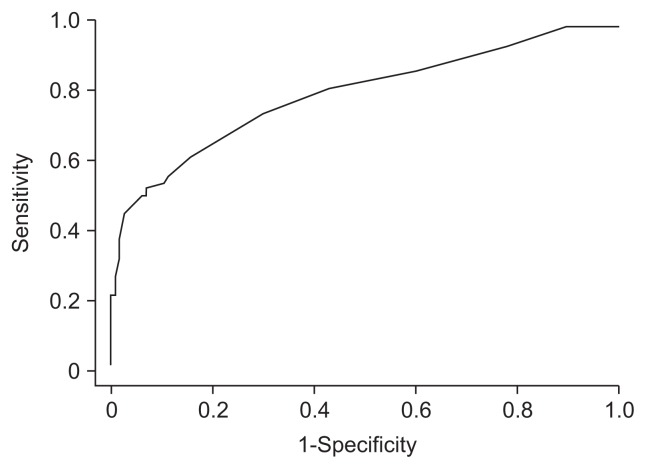
Fig. 2.
The receiver operating characteristic curve for the diagnosis of liver cirrhosis with 5805. The area under the receiver characteristic curve was 0.77. The cutoff value was Intensitym/z=5805 3.6.
3. Identification of a protein corresponding to the 5805 Da peak
The two purification steps were chosen to identify the protein or peptide 5805. First the sera of ten patients whose sera expressed high intensity 5805 Da peak were mixed to extract 5805 fraction by using the WCX Chromatography magnetic beads. The mass spectrum of the solutions was showed in Fig. 3. Then the solutions containing extracted 5805 Da fraction were further analyzed to reveal their identities using online liquid chromatography-tandem mass spectrometry (LC-MS/MS). Database searching identified the 5805 Da peak to be a C-terminal fraction of fibrinogen α-chain precursor, isoform 1 (SSSYSKQFTSSTS-YNRGDSTFESKSYKMADEAGSEADHEGTH-STKRGHAKSRPV). The one of mass spectrum of LC-MS/MS was showed in Fig. 4.
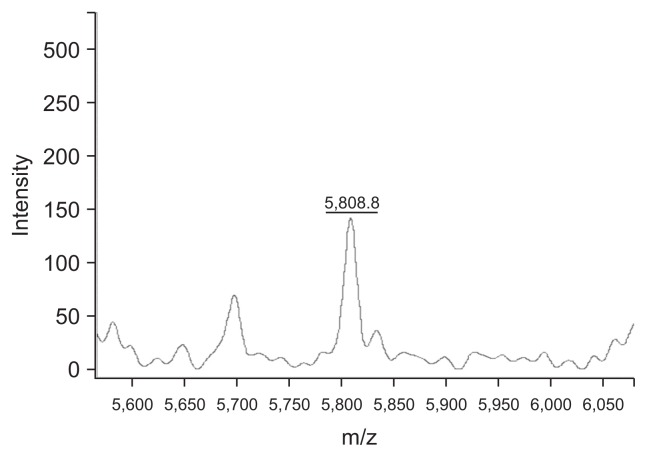
Fig. 3.
Clinprot was used to purify the 5805 peptide, and the mass spectrometry findings of the purified 5805 peptide solution are shown. The X-axis shows the m/z of the peaks, and the Y-axis shows the intensity of the peaks. The mass spectrum of the 5805 solution is also shown.
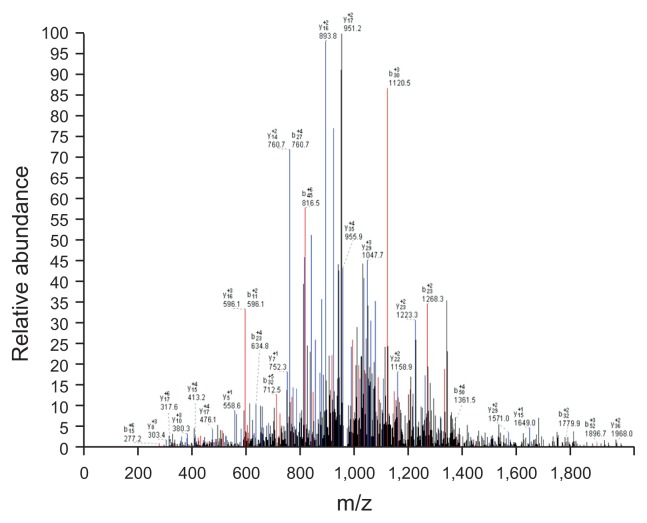
Fig. 4.
Some of the LC-MS/MS data of the extracted, highly expressed 5805 Da solution. The X-axis shows the m/z of the peaks, and the Y-axis shows the relative abundance of the b/y ion match map.
DISCUSSION
HBV infection often leads to a prolonged active viral replication, HBV DNA integration and eventually LC and its related complications.10 Hepatic fibrosis is common to many chronic liver diseases of different etiologies and could lead to cirrhosis and its related complications. Pretreatment liver biopsy has been considered as the “gold standard” for assessing the grade of liver injury and stage of liver fibrosis. Clinicians relying on liver biopsy are able to correctly diagnose the stage of fibrosis or presence of cirrhosis in 80% patients.11 However, it is associated with risks of severe complications, patient discomfort, also not suitable for regular dynamic surveillance and follow-up of disease progression.
Proteomics is a powerful strategy which can provide the global information of new proteins or peptides, disease associated targets and the process of pathogenesis by comprehensively examining different protein expression profiling between normal and pathological or drug treated samples. It has been extensively employed to investigate cancers and other diseases. The SELDI technology had been succeed in discovering tumor markers in ovary cancer, prostate cancer, and pancreatic cancer.12–14
SELDI-TOF MS was used to profile of serum for detection of the progression of chronic hepatitis C to HCC and find a peak 11735 m/z evaluated from normal to HCV-related HCC.15 As chronic hepatitis B, LC, and HCC intimidate the public health and remain the unchanged rank in high burden diseases in China. We profiled SELDI-TOF MS of the progression of chronic hepatitis B. Although there were many studies focusing on HBV-related liver disease. But most of them were interested in finding a biomarker for HCC diagnosis from patients with LC or the healthy.12–14,16
A studies profiled SELDI proteomic features of healthy people, hepatitis B-related LC patients and hepatitis B-related HCC patients and constructed an artificial neural network model to classified these three groups but not profiling the stage of chronic hepatitis B progression.17 Poon et al.18 profiled quantitatively serum of patients with chronic hepatitis B by SELDI technique and found thirty SELDI proteomic features were significantly associated with the degree of fibrosis. These thirty SELDI proteomic features were used to construct an artificial neural network model in predicting significant fibrosis (Ishak score >3) and cirrhosis (Ishak score >5) achieve a specificity of 89% and sensitivity of 89%, but none of the peaks was identified and the number of the case was small.18
In this study, we attempted to find a novel predictive bio-marker to estimate the clinic patterns ranged from diverse stages of liver fibrosis to cirrhosis and HCC based on hepatitis B using a serum proteomics approach. We luckily found a biomarker in better distinguishing liver fibrosis stage IV and cirrhosis. The SELDI peak 5805 as a diagnosis biomarker between LC (Ishak=6) and fibrosis stage IV (Ishak=5) could achieve a sensitivity of 73.3% and specificity of 85%. Meanwhile, the amino acid sequence of the 5805 peak was identified to be a fraction of isoform 1 of fibrinogen α-chain precursor (SSSYSKQFTSST-SYNRGDSTFESKSYKMA-DEAGSEADHEGTHSTKRGHAKSRPV).
Fibrinogen is composed of two sets of three polypeptide chains including α-chain, β- and γ-chain which is translated, processed, assembled in hepatic cells.19 The peripheral tissues are capable to produce the “localized” isoforms of fibrinogen as well; however, data on their structure and physiological significance remain scarce.20 Fraction of fibrinogen α-chain or isoform also were found to be evaluated in many cancers such as ovarian cancer,21 breast cancer,22 urothelial cancer,23 gastric cancer,24 and other diseases such as urinary tract infection,25 Alzheimer’s disease,26 nephritic syndrome20 even HCV-related liver disease.27 As a key organ with biological importance, the abnormal function of liver would cause a great alteration in serum protein profile undoubtedly. So the physiological significance of high level of fibrinogen fraction indicates two abnormalities in liver diseases, defection in peptide synthesis and increased protein degradation. We would postulate that the 5805 Da fraction is a complementary indicator to assess pathological initiation, development and prognosis of liver injury.
In conclusion, this study concerned the whole profiling of proteomic change of the patients with the HBV-infected disease. Though none of single protein was found to describe the exactly different stage of the liver disease, but a fraction of isoform 1 of fibrinogen α-chain precursor was identified as the biomarker for predicting worse process of liver fibrosis to LC.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34( Suppl 1):S1–S3. doi: 10.1016/S1386-6532(05)00384-7. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 3.Lok AS. Chronic hepatitis B. N Engl J Med. 2002;346:1682–1683. doi: 10.1056/NEJM200205303462202. [DOI] [PubMed] [Google Scholar]
- 4.Röcken C, Ebert MP, Roessner A. Proteomics in pathology, research and practice. Pathol Res Pract. 2004;200:69–82. doi: 10.1016/j.prp.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Tang N, Tornatore P, Weinberger SR. Current developments in SELDI affinity technology. Mass Spectrom Rev. 2004;23:34–44. doi: 10.1002/mas.10066. [DOI] [PubMed] [Google Scholar]
- 6.Poon TC. Opportunities and limitations of SELDI-TOF-MS in biomedical research: practical advices. Expert Rev Proteomics. 2007;4:51–65. doi: 10.1586/14789450.4.1.51. [DOI] [PubMed] [Google Scholar]
- 7.Zheng GX, Wang CX, Qu X, Deng XM, Deng BP, Zhang J. Establishment of serum protein pattern for screening colorectal cancer using SELDI-TOF-MS. Exp Oncol. 2006;28:282–287. [PubMed] [Google Scholar]
- 8.Bouamrani A, Ternier J, Ratel D, et al. Direct-tissue SELDI-TOF mass spectrometry analysis: a new application for clinical proteomics. Clin Chem. 2006;52:2103–2106. doi: 10.1373/clinchem.2006.070979. [DOI] [PubMed] [Google Scholar]
- 9.Engwegen JY, Gast MC, Schellens JH, Beijnen JH. Clinical proteomics: searching for better tumour markers with SELDI-TOF mass spectrometry. Trends Pharmacol Sci. 2006;27:251–259. doi: 10.1016/j.tips.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Torbenson M, Thomas DL. Occult hepatitis B. Lancet Infect Dis. 2002;2:479–486. doi: 10.1016/S1473-3099(02)00345-6. [DOI] [PubMed] [Google Scholar]
- 11.Poniachik J, Bernstein DE, Reddy KR, et al. The role of laparoscopy in the diagnosis of cirrhosis. Gastrointest Endosc. 1996;43:568–571. doi: 10.1016/S0016-5107(96)70192-X. [DOI] [PubMed] [Google Scholar]
- 12.Vlahou A, Schellhammer PF, Mendrinos S, et al. Development of a novel proteomic approach for the detection of transitional cell carcinoma of the bladder in urine. Am J Pathol. 2001;158:1491–1502. doi: 10.1016/S0002-9440(10)64100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam BL, Qu Y, Davis JW, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- 14.Rosty C, Christa L, Kuzdzal S, et al. Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a bio-marker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 2002;62:1868–1875. [PubMed] [Google Scholar]
- 15.Schwegler EE, Cazares L, Steel LF, et al. SELDI-TOF MS profiling of serum for detection of the progression of chronic hepatitis C to hepatocellular carcinoma. Hepatology. 2005;41:634–642. doi: 10.1002/hep.20577. [DOI] [PubMed] [Google Scholar]
- 16.Geng X, Wang F, Li YG, Zhu GP, Zhang WM. SELDI-TOF MS proteinchip technology for screening of serum markers of HBV-induced hepatocellular carcinoma. J Exp Clin Cancer Res. 2007;26:505–508. [PubMed] [Google Scholar]
- 17.Cui J, Kang X, Dai Z, et al. Prediction of chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma by SELDI-based serum decision tree classification. J Cancer Res Clin Oncol. 2007;133:825–834. doi: 10.1007/s00432-007-0224-y. [DOI] [PubMed] [Google Scholar]
- 18.Poon TC, Hui AY, Chan HL, et al. Prediction of liver fibrosis and cirrhosis in chronic hepatitis B infection by serum proteomic fingerprinting: a pilot study. Clin Chem. 2005;51:328–335. doi: 10.1373/clinchem.2004.041764. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, Cao Z, Chung DW, Davie EW. The role of betagamma and alphagamma complexes in the assembly of human fibrinogen. J Biol Chem. 1996;271:27942–27947. doi: 10.1074/jbc.271.44.27942. [DOI] [PubMed] [Google Scholar]
- 20.Picken MM, Linke RP. Nephrotic syndrome due to an amyloidogenic mutation in fibrinogen A alpha chain. J Am Soc Nephrol. 2009;20:1681–1685. doi: 10.1681/ASN.2008070813. [DOI] [PubMed] [Google Scholar]
- 21.Ogata Y, Heppelmann CJ, Charlesworth MC, et al. Elevated levels of phosphorylated fibrinogen-alpha-isoforms and differential expression of other post-translationally modified proteins in the plasma of ovarian cancer patients. J Proteome Res. 2006;5:3318–3325. doi: 10.1021/pr060344+. [DOI] [PubMed] [Google Scholar]
- 22.Gast MC, Van Gils CH, Wessels LF, et al. Serum protein profiling for diagnosis of breast cancer using SELDI-TOF MS. Oncol Rep. 2009;22:205–213. doi: 10.3892/or_00000426. [DOI] [PubMed] [Google Scholar]
- 23.Theodorescu D, Wittke S, Ross MM, et al. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 24.Su Y, Shen J, Qian H, et al. Diagnosis of gastric cancer using decision tree classification of mass spectral data. Cancer Sci. 2007;98:37–43. doi: 10.1111/j.1349-7006.2006.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pacchiarotta T, Hensbergen PJ, Wuhrer M, et al. Fibrinogen alpha chain O-glycopeptides as possible markers of urinary tract infection. J Proteomics. 2012;75:1067–1073. doi: 10.1016/j.jprot.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Vafadar-Isfahani B, Ball G, Coveney C, et al. Identification of SPARC-like 1 protein as part of a biomarker panel for Alzheimer’s disease in cerebrospinal fluid. J Alzheimers Dis. 2012;28:625–636. doi: 10.3233/JAD-2011-111505. [DOI] [PubMed] [Google Scholar]
- 27.Sogawa K, Noda K, Umemura H, et al. Serum fibrinogen alpha C-chain 5.9 kDa fragment as a biomarker for early detection of hepatic fibrosis related to hepatitis C virus. Proteomics Clin Appl. 2013;7:424–431. doi: 10.1002/prca.201200094. [DOI] [PubMed] [Google Scholar]