Fig. 3.
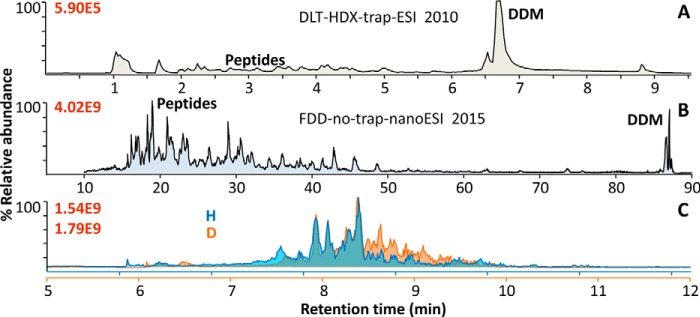
At much higher peptide ion sensitivity, the 2015 FDD-no-trap-nESI method proved DDM can be a superior tool for broad nESI-based membrane proteomics. Representative HPLC MS traces of (A) 2010 DLT-HDX-trap-ESI of hGPCR apo β2AR, typical to the data for (6), B, FDD-no-trap-nESI of hGABAAR, and (C) independent digests of H/D versions of hGABAAR using FDD-no-trap-nESI's HDX module showing good reproducibility. All spectra were acquired with orbitrap analyzers in Thermo Exactive (A), Q-Exactive (B), or LTQ-orbitrap XL (C) mass spectrometer. Experiments used similar concentrations of protein and detergent, but A loaded several-fold more total sample to HPLC than B and C. Figure was adapted from (7).
