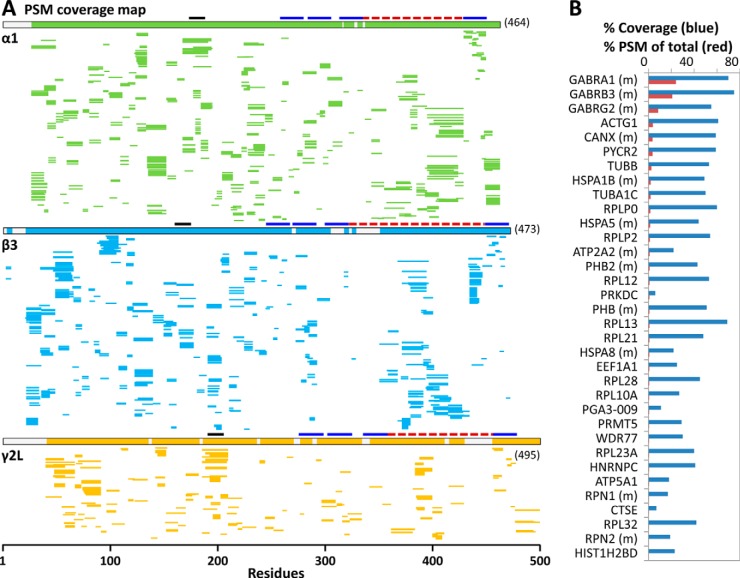
Fig. 4.
Protein sequence coverage identified from direct FDD-pepsin digestion and analysis of DDM/CHS-enriched hGABAAR solution, by searching against. A, the target sequences of (α1)2(β3)2(γ2L)1 GABAAR (SEQUEST in Proteome Discoverer 1. 3), or (B) the original human proteome (May 23, 2013 Ensembl, SEQUEST-HT in Proteome Discoverer 1.4), both at no protease specificity and filtered to peptide FDR <1% by Percolator. Each thin line represents one PSM (apple green, α1 subunit; blue, β3; orange, γ2L); bold lines indicate domains (dark blue, TM; red, intracellular loop; black, extracellular Cys-loop). B, shows the % sequence coverage (blue bar) for the top 34 proteins (ranked by PSMs and unique peptides), and their percent PSM (red bar) of the total PSMs of all proteins identified by three or more valid unique peptides; (m) designates membrane residence, as annotated in human proteome database. N-terminal signaling peptide sequences were not removed. A was adapted from (7). These proteins were mainly cell-originated survivors of two affinity screenings by membrane isolation and by buffer washing during anti-FLAG enrichment, not contaminant from processing.