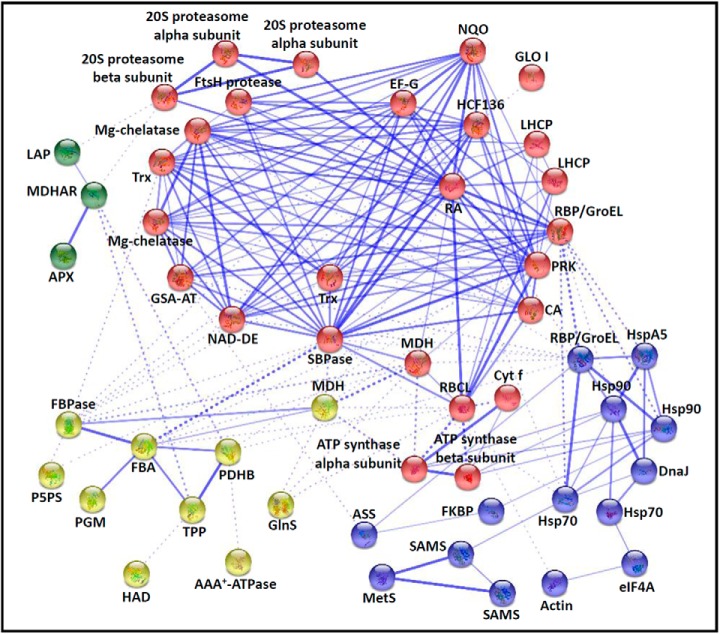
Fig. 7.
The protein–protein interaction (PPI) network in O. cinnamomea L. var. asiatica spores revealed by STRING analysis. A total of 100 differentially expressed proteins represented by 53 unique homologous proteins from Arabidopsis are shown in PPI network. Four main groups are indicated in different colors. The PPI network is shown in the confidence view generated by STRING database. Stronger associations are represented by thicker lines. APX, ascorbate peroxidase; ASS, argininosuccinate synthase; CA, carbonic anhydrase; DnaJ, chaperone DnaJ; EF-G, translation elongation factor G; eIF4A, eukaryotic initiation factor 4A; FBA, fructose-1,6-bisphosphate aldolase; FBPase, fructose-1,6-bisphosphatase; FKBP, FK506 binding protein-type peptidyl-prolyl cis-trans isomerase; GlnS, glutamine synthetase; GLO I, glyoxalase I; GroEL, GroEL like type I chaperonin; GSA-AT, glutamate-1-semialdehyde aminotransferase; HAD, beta-hydroxyacyl-acyl carrier protein-dehydratase; HCF136, photosystem II stability/assembly factor HCF136; Hsp 70/90, heat shock protein 70/90; HSPA5, heat shock 70 protein 5; LAP, leucine aminopeptidase; LHCP, light harvesting chlorophyll a/b binding protein, photosystem II; MDH, malate dehydrogenase; MDHAR, monodehydroascorbate reductase; MetS, methionine synthase; Mg-chelatase, Mg-protoporphyrin IX chelatase; NAD-DE, NAD-dependent epimerase/dehydratase; NQO, NADH: quinone oxidoreductase-like protein; P5PS, pyridoxal 5′-phosphate synthase; PDHB, pyruvate dehydrogenase E1 component beta subunit; Cyt f, cytochrome f; PGM, phosphoglucomutase; PRK, phosphoribulokinase; RA, ribulose-1,5-bisphosphate carboxylase activase; RBCL, ribulose-1,5-bisphosphate carboxylase large subunit; RBP, ribulose-1,5-bisphosphate carboxylase large subunit-binding protein; SAMS, S-adenosylmethionine synthetase; SBPase, sedoheptulose-1,7-bisphosphatase; TPP, thiamine pyrophosphate; Trx, thioredoxin.