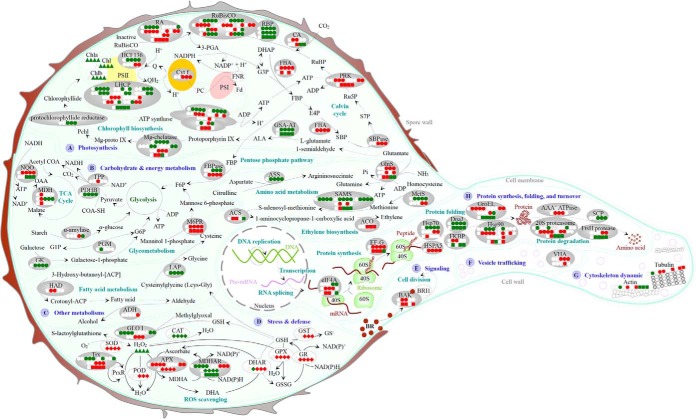
Fig. 9.
Schematic presentation of the O. cinnamomea L. var. asiatica spore germination mechanisms. The identified proteins were integrated into subcellular pathways. A, Photosynthesis; B, Carbohydrate and energy metabolism; C, Other metabolisms; D, Stress and defense; E, Signaling; F, Vesicle trafficking; G, Cytoskeleton dynamic; H, Protein synthesis, folding, and turnover. Protein expression patterns, gene expression patterns, enzyme activities, and substrate contents are marked with circles, squares, diamonds, and triangles in white (unchanged), red (increased), and green (decreased), respectively. The solid line indicates single-step reaction, and the dashed line indicates multistep reaction. 3-PGA, 3-phosphoglyceric acid; 40S, eukaryotic small ribosomal subunit; 60S, eukaryotic large ribosomal subunit; Acetyl COA, acetyl coenzyme A; ACP, acyl carrier protein; ADH, alcohol dehydrogenase; ALA, 5-aminolevulinic acid; ANK, ankyrin-repeat containing protein; APX, ascorbate peroxidase; ASS, argininosuccinate synthase; BAK, brassinosteroid LRR receptor kinase; BR, brassinosteroid; BRI1, brassinosteroid-insensitive 1; CA, carbonic anhydrase; CaM, calmodulin; CAT, catalase; Chl, chlorophyll; COA-SH, coenzyme A; Cyt f, cytochrome f; DHA, dehydroascorbic acid; DHAP, dihydroxyacetone phosphate; DHAR, dehydroascorbate reductase; E4P, erythrose 4-phosphate; EF-G, translation elongation factor G; eIF4A, eukaryotic initiation factor 4A; F6P, fructose 1,6-diphosphate; FBA, fructose-1,6-bisphosphate aldolase; FBP, fructose 1,6-bisphosphate; FBPase, fructose-1,6-bisphosphatase; Fd, ferredoxin; FKBP, FK506 binding protein-type peptidyl-prolyl cis-trans isomerase; FNR, ferredoxin-NADP+ reductase; G1P, glucose-1-phosphate; G3P, glyceraldehyde 3-phosphate; G6P, glucose-6-phosphate; GK, galactose kinase; GLO I, glyoxalase I; GPX, glutathione peroxidase; GR, glutathione reductase; GroEL, GroEL like type I chaperonin; GlnS, glutamine synthetase; GSA-AT, glutamate-1-semialdehyde aminotransferase; GSH, glutathione; GSSG, oxidized glutathione; GST, glutathione S-transferase; HAD, beta-hydroxyacyl-acyl carrier protein-dehydratase; Hsp, heat shock protein; HSPA5, heat shock 70 protein 5; LAP, leucine aminopeptidase; LHCP, light harvesting chlorophyll a/b binding protein; M6PR, mannose-6-phosphate reductase; MDH, malate dehydrogenase; MDHA, monodehydroascorbate; MDHAR, monodehydroascorbate reductase; Mg-chelatase, Mg-protoporphyrin IX chelatase; Mg-proto IX, Mg-protoporphyrin IX; MetS, methionine synthase; NQO, NADH-quinone oxidoreductase; OAA, oxaloacetate; PC, plastocyanin; Pchl, protochlorophyllide; PDHB, pyruvate dehydrogenase E1 component beta subunit; PGM, phosphoglucomutase; POD, peroxidase; PRK, phosphoribulokinase; PrxR, peroxiredoxin; PSI, photosystem I; PSII, photosystem II; Q, quinone; QH2, hydroquinone; RA, ribulose-1,5-bisphosphate carboxylase activase; RBP, ribulose-1,5-bisphosphate carboxylase large subunit-binding protein; ROS, reactive oxygen species; RuBisCO, ribulose-1,5-bisphosphate carboxylase; Ru5P, ribulose-5-phosphate; RuBP, ribulose-1,5-bisphosphate; S7P, sedoheptulose 7-phosphate; SAMS, S-adenosylmethionine synthetase; SBP, sedoheptulose 1,7-bisphosphate; SBPase, sedoheptulose-1,7-bisphosphatase; SCP, serine carboxypeptidase; SOD, superoxide dismutase; TCA, tricarboxylic acid; TPP, thiamine pyrophosphate; Trx, thioredoxin; VHA, vacuolar H+-ATPase.