Table I. Differentially expressed proteins during Osmunda cinnamomea L. var. asiatica. spore germination.
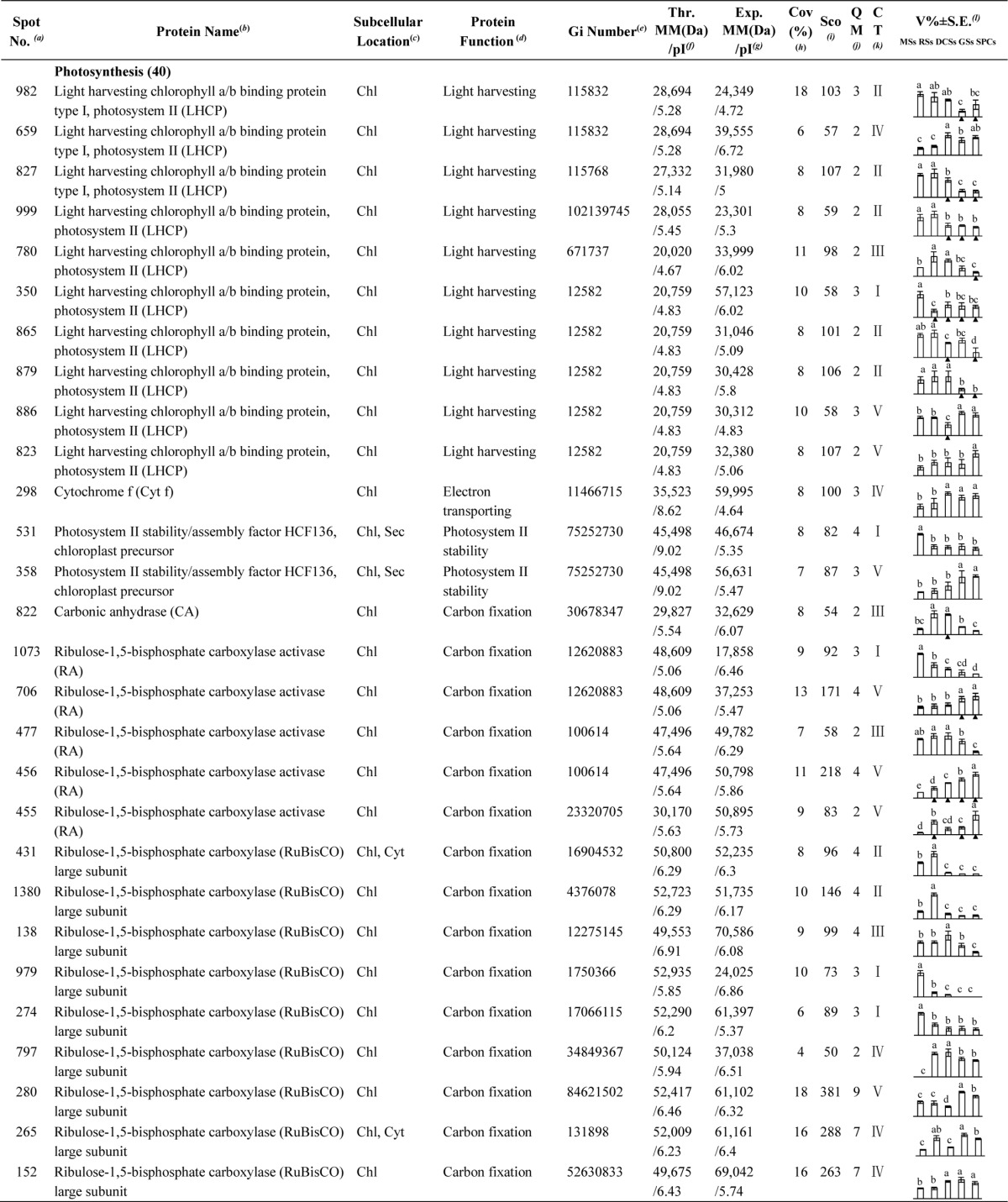
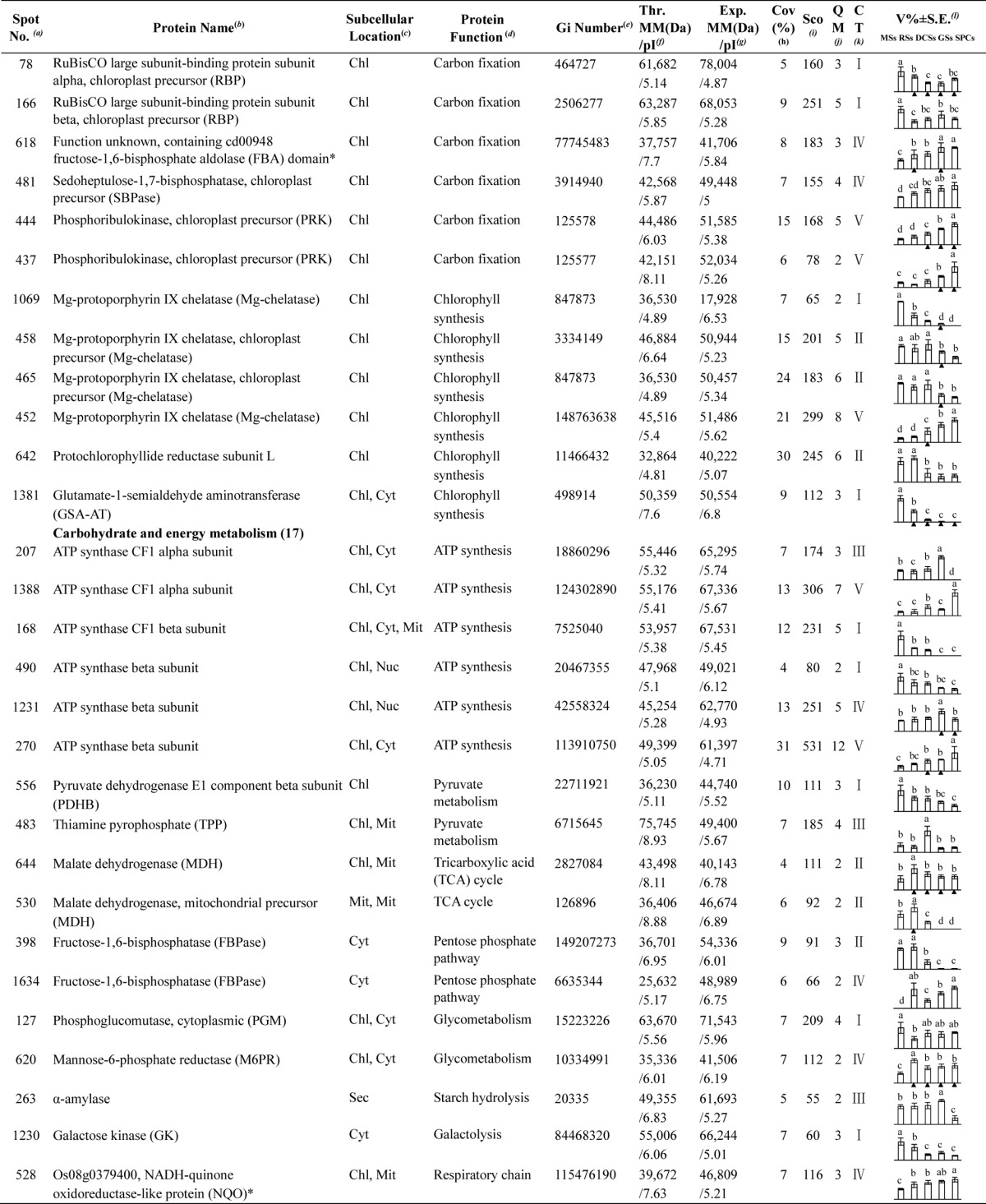
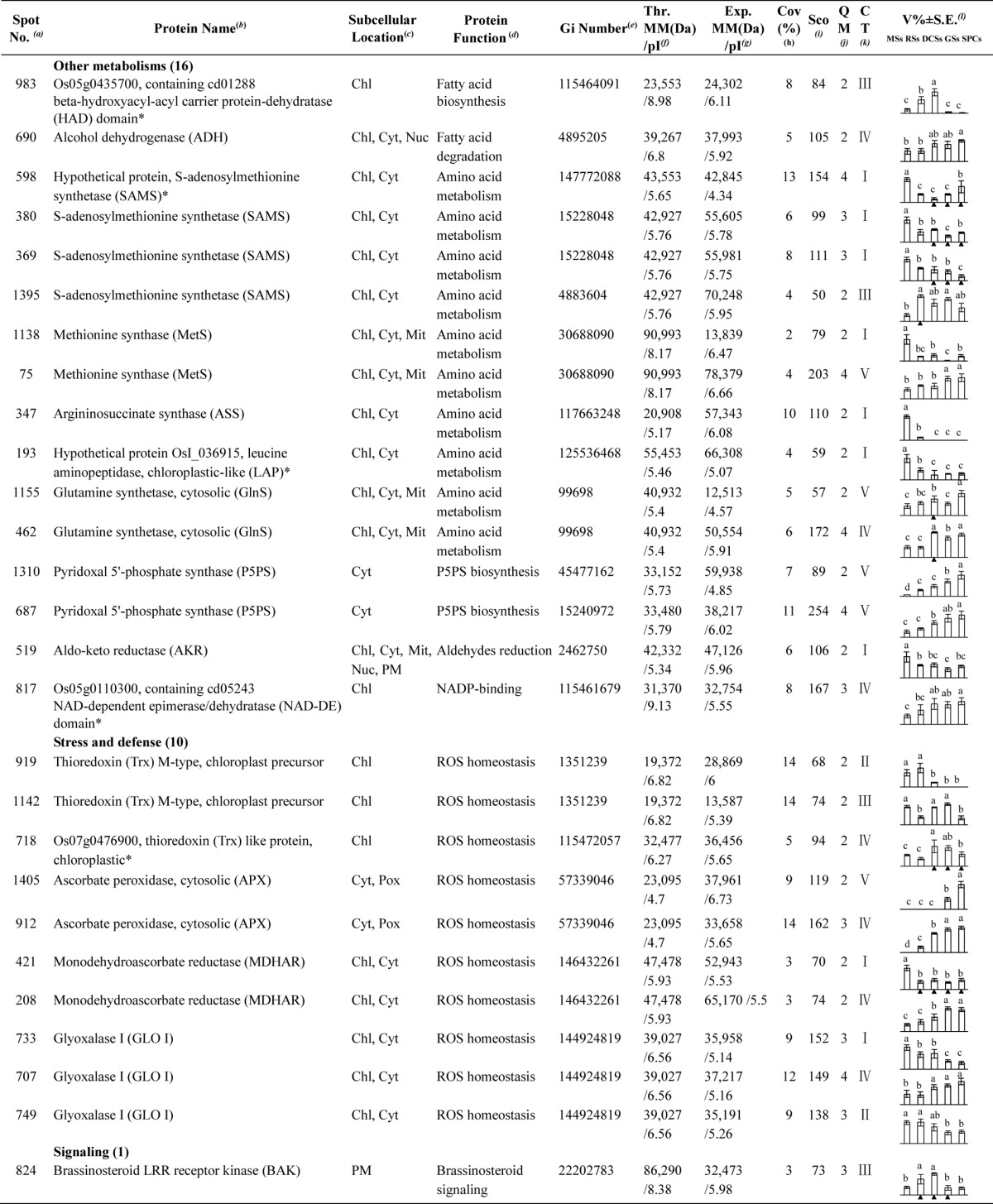
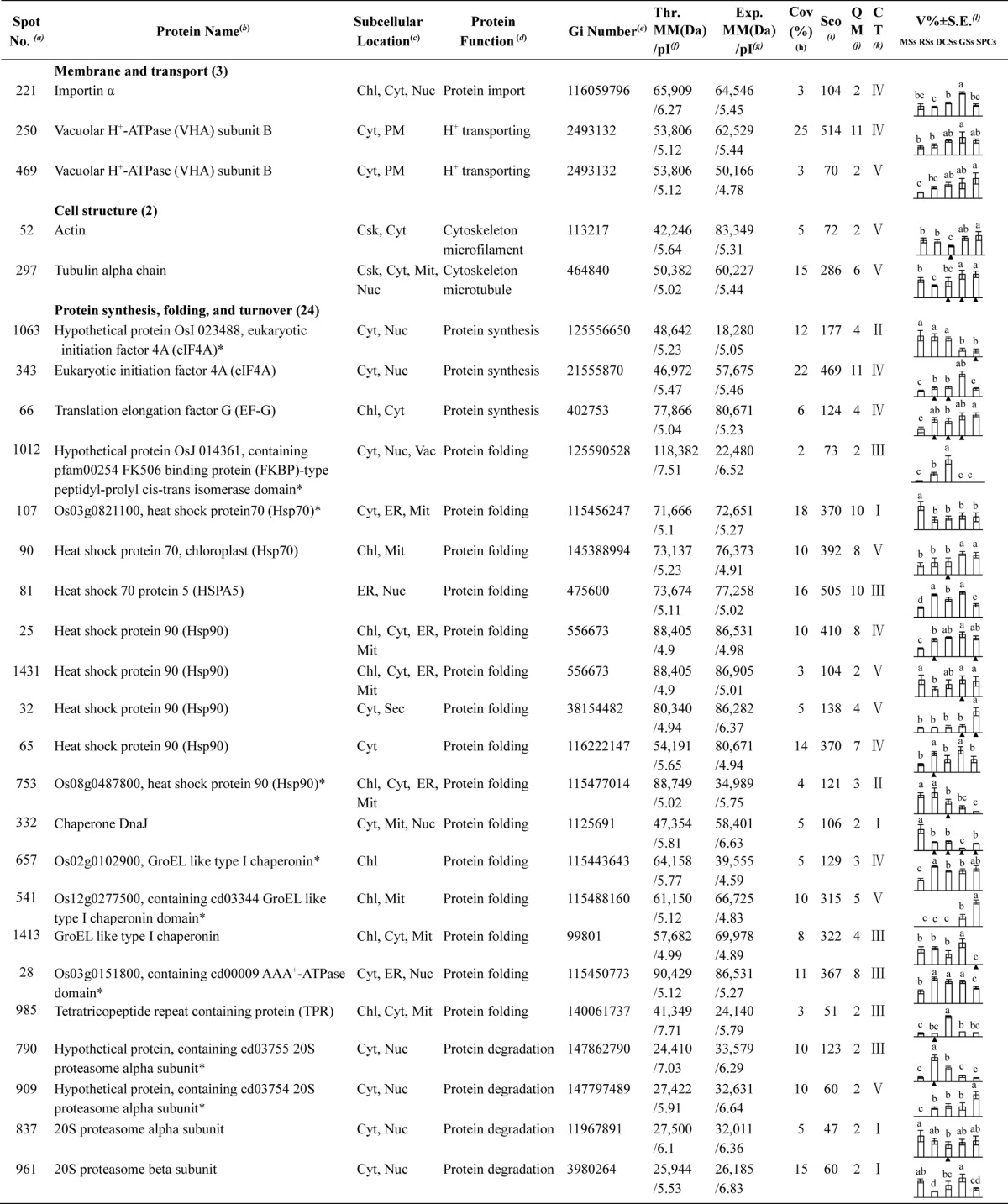
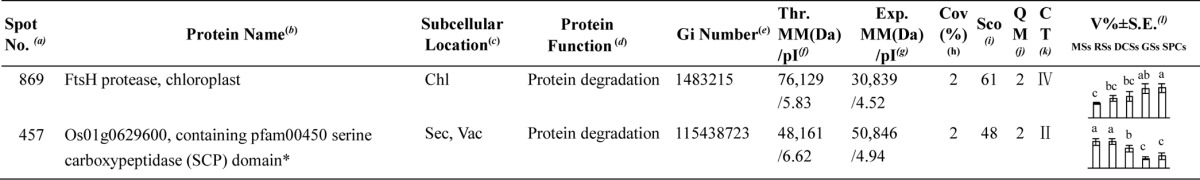
a Assigned spot number as indicated in Fig. 5.
b The name and functional category of the proteins identified by ESI-Q-TOF MS. Protein names marked with an asterisk (*) have been edited by us depending on functional domain searching and similarity comparison according to the Gene Ontology criteria.
c Protein subcellular localization predicted by softwares (YLoc, LocTree3, Plant-mPLoc, ngLOC, and TargetP). Chl, chloroplast; Csk, cytoskeleton; Cyt, cytoplasm; ER, endoplasmic reticulum; PM, plasma membrane; Mit, mitochondria; Nuc, nucleus; Pox, peroxisome; Sec, secreted; Vac, vacuole.
d The molecular function of the identified proteins.
e Database accession number from NCBInr.
f,gTheoretical (f) and experimental (g) molecular mass (Da) and pI of identified proteins. Experimental values were calculated using ImageMaster 2D software. Theoretical values were retrieved from the protein database.
hThe amino acid sequence coverage for the identified proteins.
iThe Mascot score obtained after searching against the NCBInr database.
jThe number of unique peptides identified for each protein.
kThe cluster types of identified proteins obtained from the GeneCluster software (version 2.0).
lThe mean values of protein spot volumes relative to total volume of all the spots. The black triangle (▴) under the column of protein abundance indicates that the proteins showed similar trends with their homologous genes (Please refer to Fig. 8). Five spore germination stages (MSs, RSs, DCSs, GSs, and SPCs) were performed. MSs, mature spores; RSs, rehydrated spores; DCSs, double-celled spores; GSs, germinated spores; SPCs, spores with protonemal cells. Error bar indicates ± standard error (S.E.). Letters indicate statistically significant differences (p < 0.05) among five stages of spore germination as determined by one-way ANOVA.
