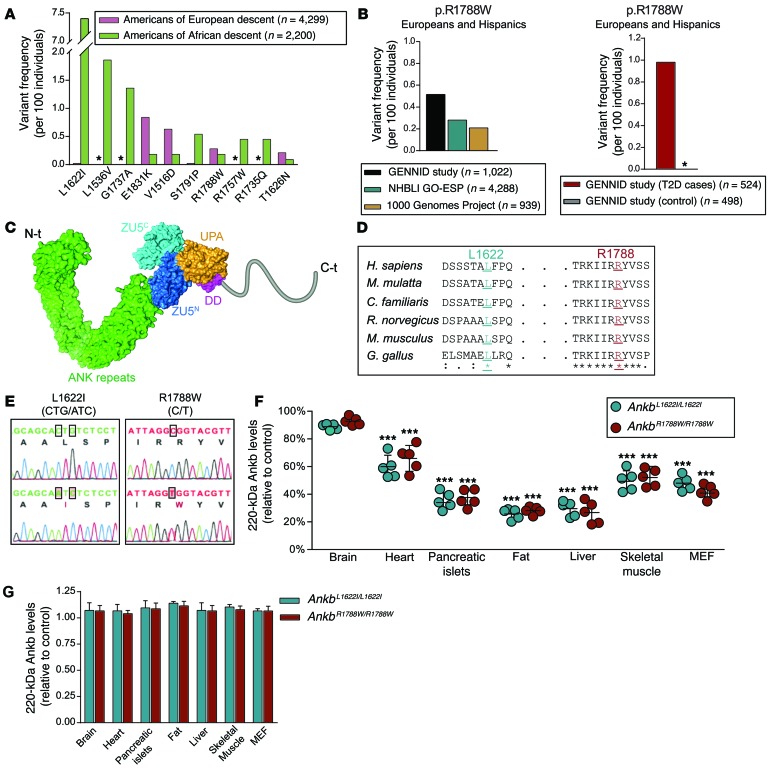
Figure 1. Human AnkB variants cause tissue-specific AnkB reduction.
(A) Variant frequency (per 100 individuals) for the most common ANKB variants in North Americans of European and African descent. Allele frequencies were compiled from the NHLBI GO Exome Sequencing Project. Asterisks indicate undetected variants. (B) Frequency of the p.R1788W ANKB variant in European Americans and Hispanics from the GENNID cohort of noninsulin-dependent diabetic patients (control and T2D cases) and from individuals of the same ethnicity sequenced as part of the entire GENNID cohort, the NHLBI GO Exome Sequencing Project, and the 1000 Genomes Project. Asterisks indicate undetected variants. (C) Model of AnkB structure, with domains indicated, showing the localization of L1622 and R1788 sites within AnkB’s unstructured C-terminal domain. (D) Evolutionary conservation of L1622 and R1788 sites. (E) Sequencing chromatograms of brain cDNA from Ankb+/+ and Ankb knockin mice. (F) Quantitative analysis of AnkB protein levels for the indicated tissues as a percentage of control (Ankb+/+). (G) Quantification of 220-kDa Ankb transcript levels by qPCR. GAPDH was used for normalization of mRNA and protein levels. Data represent mean ± SEM (n = 5 males, 10-weeks-old). Results are representative of 3 independent experiments. ***P < 0.001, 2-tailed t test.