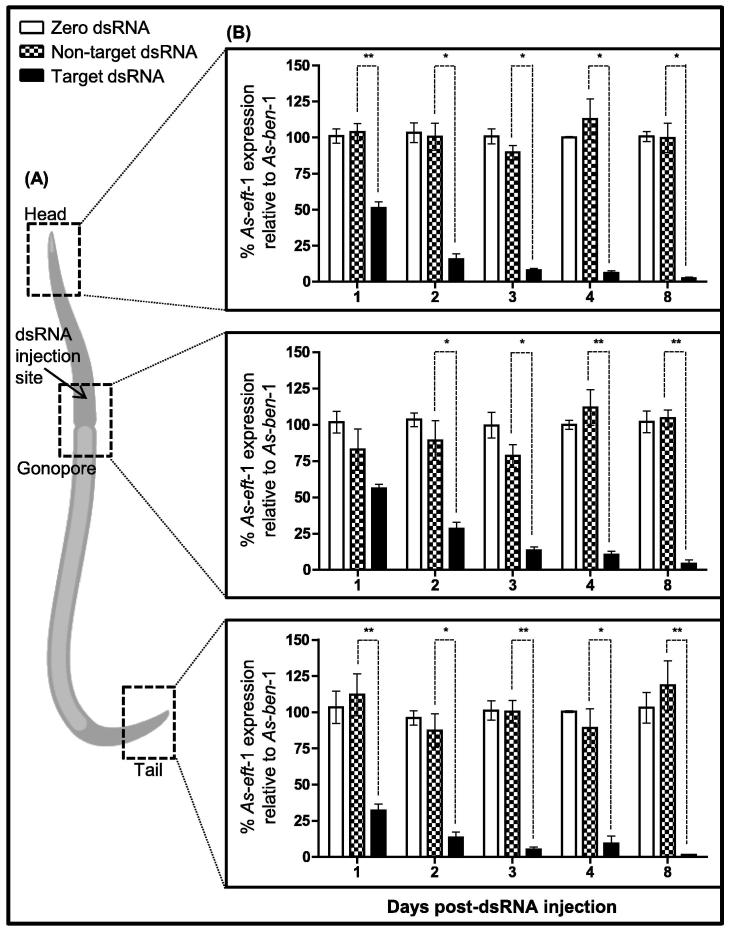
Fig. 1.
A novel adult Ascaris suum RNA interference (RNAi) trigger delivery approach induces a robust systemic RNAi response in distinct tissue regions. (A) Novel RNAi trigger delivery approach: double-stranded RNA (dsRNA) was injected anterior to the gonopore and transcript levels were assessed in head, gonopore and tail tissue segments. (B) A. suum translational elongation factor 1a (As-eft-1) target transcript knockdown was significant (>95% by day 8, P < 0.05), and persistent (up to 8 days) in tissues remote from the RNAi trigger delivery site. Head region: As-eft-1 knockdown in target (As-eft-1) dsRNA treated worms compared with zero dsRNA controls is 48.8%, 84.4%, 92%, 94.1% and 97.6% at 1, 2, 3, 4 and 8 days post-injection, respectively (n = 6 for all time points except day 4 where n = 4); gonopore region: As-eft-1 knockdown in target dsRNA treated worms compared with zero dsRNA controls is 64%, 71.7%, 86.5%, 89.6% and 95.7% at 1, 2, 3, 4 and 8 days post-injection, respectively (n = 6); tail region: As-eft-1 knockdown in target dsRNA treated worms compared with zero dsRNA controls is 67.8%, 86.7%, 94.8%, 90.8% and 98.5% at 1, 2, 3, 4 and 8 days post-injection, respectively (n = 6 for all time points except day 4 where n = 4). Note that A. suum elongation factor 1b (As-eft-2) knockdown (head: 87.7 ± 2.8% (n = 6; P < 0.05, target dsRNA versus non-target dsRNA); gonopore: 81.1 ± 10.4% (n = 6; P < 0.05, target dsRNA versus non-target dsRNA); tail: 96.7 ± 1.2% (n = 6; P < 0.05, target dsRNA versus non-target dsRNA)) was assessed in As-eft-1-dsRNA treated worms compared with zero dsRNA control worms, 8 days post-injection (graph not shown); error bars represent S.E.M.; *P < 0.05, **P < 0.01. Methods were as follows: (i) dsRNA synthesis. Target and control T7-labelled PCR products (target and control GenBank accession numbers, sense (S) and antisense (AS) primers (5′-3′), and amplicon sizes were: As-eft-1 (Elongation factor 1a; accession No. BK006061), S 5′-GAGGCTCTGTCGCTTCTGAC-3′, AS 5′-TTGAGGAACTTCGGGTTGTC-3′ (219 bp); As-eft-2 (Elongation factor 1b; accession No. JI172981) S 5′-AATGAACCACCCGGGACAGA-3′, AS 5′-TCAACGCACAGCGGTTTTGT-3′ (201 bp); As-gmpr (GMP reductase; accession No. JO467845), S 5′-ATCTTTGCGGGCAATGTCGT-3′, AS 5′-TGCCCGTTGAGACCATGAGA-3′ (185 bp); As-tnc-1 (troponin C; accession No. JO472030), S 5′-CGACGAGTTTTGCGCTCTTG-3′, AS 5′-TCCACAGCCGCTTCTAGCTG-3′ (192 bp); As-rab-3 (synaptic vesicle protein; accession No. JI178604), S 5′-CCGATCAGCTCGGTTTGGAG-3′, AS 5′-TGCACTGTTGTGATGGTGGTTT-3′ (195 bp); As-hb-1 (haemoglobin; accession No. AAA29374), S 5′-TCTTTGCGCCACCTACGATG-3′, AS 5′-ACTCCCTGCCGATTTCATGC-3′ (197 bp); As-unc-29 (nicotinic acetylcholine receptor subunit; accession No. ADY42984), S 5′-ACTTTCGGGGACCGACCACT-3′, AS 5′-GTGATGTGCGGACGGATTCA-3′ (209 bp); As-unc-38 (nicotinic acetylcholine receptor subunit; accession No. ERG87157), S 5′-TGGTGGTGTCAGCGTGCTTT-3′, AS 5′-TGCTCATCGAATGGAAACCATC-3′ (204 bp); non-target control dsRNA (Neomycin Phosphotransferase; accession No. U55762), S 5′-GGTGGAGAGGCTATTCGGCT-3′, AS 5′-CCTTCCCGCTTCAGTGACAA-3′ (223 bp); note that dsRNA T7 template primers had a T7 promoter (5′-TAATACGACTCACTATAGGG-3′) appended to the 5′ end) with sense and antisense polarity were amplified (cycling conditions: 95 °C × 5 min, 40 × (95 °C × 30 s, 60 °C × 30 s, 72 °C × 2 min), 72 °C × 10 min) from sequence-verified cDNA templates (GATC Biotech, Germany, http://www.gatc-biotech.com), and used to synthesise dsRNA with either the MEGAshortscript™ Kit (Life Technologies, USA), or T7 RiboMAX™ Express RNAi kit (Promega, USA) according to manufacturer’s instructions. dsRNA was eluted in Ascaris saline (125 mM CH3COONa, 24.5 mM KCl, 4 mM NaCl, 11.8 mM CaCl2, 9.8 mM MgCl2, 5 mM MOPS buffer, pH 6.8), and visualised on a 1.2% agarose gel to confirm integrity. Concentration and purity were assessed using a Nanodrop ND-1000-A spectrophotometer (Labtech, UK). (ii) dsRNA delivery and tissue dissection. Adult female A. suum (⩾15 cm) were injected (26 gauge needle) with 100 μl of dsRNA (200 ng/μl) or Ascaris saline (zero dsRNA control), into the pseudocoelomic cavity (1 cm anterior to the gonopore on the ventral side of the worm). dsRNA and Ascaris saline was supplemented with 1% green food dye to facilitate visualisation of pseudocoelomic delivery. Ascaris suum were maintained in Ascaris Ringer Solution (13.14 mM NaCl, 9.47 mM CaCl2, 7.83 mM MgCl2, 12.09 mM C4H11NO3/Tris, 99.96 mM NaC2H3O2, 19.64 mM KCl, pH 7.8) for 1–8 days post-injection after which tissue sections from the head (1.5 cm segment), tail (1.5 cm segment) and gonopore regions (1 cm segment) were dissected, snap-frozen in liquid nitrogen and stored at −80 °C until use. Where relevant, muscle bag cells were dissected as a tissue layer from a body wall flap (1 cm long; including both dorsal and ventral tissue), that was excised ∼2 cm anterior to the gonopore. Post-dissection, muscle bag cell layers were submerged in autoclaved Ascaris perienteric fluid (APF: 23 mM NaCl, 110 mM NaC2H3O2, 24 mM KCl, 6 mM CaCl2, 5 mM MgCl2, 11 mM C6H12O6, 5 mM C8H18N2O4S, pH 7.6). Sterile scissors were used to snip the arm region of the muscle tissue and release the layer of muscle bag cells from the rest of the body wall. Note that both ventral and dorsal muscle bag cell layers were dissected from the same body wall flap and processed together. (iii) Post-RNAi transcript analysis. Individual tissue segments were ground to a fine powder whilst frozen using a mortar and pestle. Total RNA was extracted using TRIzol® Reagent (Life Technologies), treated with DNase I (Ambion TURBO DNase, Life Technologies), standardised to 1800 ng/μl, and used for cDNA synthesis (Applied Biosystems High Capacity RNA-to-cDNA reverse transcription kit (Life Technologies) according to manufacturer’s instructions). Target and reference gene (As-ben-1) transcripts were amplified from each cDNA in triplicate by quantitative real-time PCR (qPCR) using a Qiagen Rotor-Gene 5-plex HRM, and FastStart SYBR Green Master (Roche, Switzerland) (target and reference gene S- and AS primers, and amplicon sizes were as follows: As-eft-1, S 5′-ATTCTCCGAGACTCGCTTCA-3′, AS 5′-GAGATCGGCACGAAAGCTAC-3′ (99 bp); As-eft-2, S 5′-GTCCCACCACAGAGGCCAAC-3′, AS 5′-CACGACCCACTGGCACTGTC-3′ (94 bp); As-gmpr, S 5′-GAGTGGAAGGCTTTCGTGCAA-3′, AS 5′-CCGATGTCGATATTCCCGATG-3′ (85 bp); As-tnc-1, S 5′-TCGATGGATCGCAAATCGAA-3′, AS 5′-TGGGTGGCCATGATGTAACC-3′ (82 bp); As-rab-3, S 5′-CGCGGGGATAAACGTGTCA-3′, AS 5′-GAAGCCCATTGCACCACGAT-3′ (105 bp); As-hb-1, S 5′-GCTTTCAAGGACCGCGAGAA-3′, AS 5′-GCAGATGACAGGCGAGCAGA-3′ (97 bp); As-unc-29, S 5′-CCCCACTGATTCAGGCGAAA-3′, AS 5′-TGGGTGGCAGGATCTTCGAT-3′ (95 bp); As-unc-38, S 5′-GGCCACTTGCTGATACCGATG-3′, AS 5′-GGCTTACGCGATGTTCGTGA-3′ (102 bp); As-ben-1 (beta-tubulin; GenBank accession number FE913811), S 5′-CCCACATACGGAGACCTCAACC-3′, AS 5′-CCAATTTGCGCAAGTCTGCAT-3′ (103 bp); cycling conditions: 9 °C × 10 min, 40 × (95 °C × 15 s, 60 °C × 15 5 s, 72 °C × 30 s), 72 °C x10 min). PCR efficiencies were calculated using Real-time PCR Miner (http://www.miner.ewindup.info/index.htm) and used for relative quantification of target gene transcript levels by the augmented comparative ΔΔCt method (Pfaffl, 2001). Changes in target gene transcript abundance were analysed using the non-parametric Kurskal–Wallis test, and the Dunn’s post-test (to assess the significance of the zero dsRNA and target dsRNA treatments against the non-target dsRNA, separately; note that we did not detect a significant difference between zero dsRNA and non-target dsRNA controls in any dataset) with GraphPad PRISM Version 5 (GraphPad Software, Inc., USA).