Abstract
A microfluidic platform is presented which fully automates all incubation steps of a three-stage, multiplexed magnetic bead immunoassay, such as the Luminex® xMAP technology. Magnetic actuation is used to transfer the microbeads between co-infused adjacent laminar flow streams to transport the beads into and out of incubation and wash solutions, with extended incubation channels to allow sufficient bead incubation times (1–30 min, commonly 5 min per stage) to enable high-sensitivity. The serial incubation steps of the immunoassay are completed in succession within the device with no operator interaction, and the continuous flow operation with magnetic bead transfer defines the incubation sequencing requiring no external fluidic controls beyond syringe pump infusion. The binding kinetics of the assay is empirically characterized to determine the required incubation times for specific assay sensitivities in the range 1 pg/ml to 100 ng/ml. By using a Luminex® xMAP duplex assay, concurrent detection of IL-6 and TNF-α was demonstrated on-chip with a detection range 10 pg/ml to 1 ng/ml. This technology enables rapid automation of magnetic microbead assays, and has the potential to perform continuous concentration monitoring.
Keywords: Immunoassay, Microfluidic, Biosensor, Magnetic, Fluorocytometry
1 Introduction
Many benefits have been promised from the application of microfluidics to clinical diagnostics and life sciences research, such as low sample volume requirements, parallelization, and portability. An additional advantage which has been somewhat underreported in the literature thus far is automation. Automation often requires a large amount of systems integration and is thus difficult to demonstrate at the laboratory research level. The state of the art in assay automation is through the use of robotics to perform traditional assays. This technology has been able to increase labor efficiency, improve the reliability of results, and enable high throughput with fast operation and the use of high count well plates. However, these systems are impractical in applications where a small number of samples must be run on an automated platform, and where a short assay turnaround time is desired with only one sample, such as is the case for many clinical tests. Furthermore, the use of robotics involves significant maintenance, extensive operator training, and generally requires a large capital investment, all of which hinder its use in many clinical applications as well as certain laboratory applications. By employing novel operating modes, microfluidic devices have the ability to provide assay automation for these applications while alleviating many of the problems associated with current automation methods. These types of microfluidic devices which automate assays without complex mechanical systems may be described as intrinsic design assay automation. In these systems, the automation is inherent to the microfluidic design, and the assay is driven solely by the flow of reagents into the device without external controls. Intrinsic design automation with microfluidics offers numerous benefits such as lower overall system cost, minimal sample and reagent volume requirements, and reduced operator training requirements.
In this work, we present a microfluidic intrinsic design assay automation device which performs the incubation steps of magnetic microbead immunoassays. The use of microbeads as an immobilization surface for binding assays offers unique advantages over surface immobilization for microfluidic designs due to the ease of renewability. Microbead surfaces can easily be batch-functionalized prior to the assay, infused into a closed channel, and manipulated within the microfluidic chip. There are examples of single use, single sample, non-renewable microfluidic assay designs using microbeads. In these systems, the use of microbeads increases the binding surface area relative to the channel volume when compared to functionalization of only the microchannel surfaces. These devices use passive retention systems to retain the infused microbeads while assay reagents flow through. Retention can be accomplished with physical barriers such as steps and posts (Andersson et al. 2000; Oleschuk et al. 2000; Sato et al. 2000, 2004; Jeong et al. 2008), wells (Thompson et al. 2010), or gels (Breadmore et al. 2003), or by other forces such as magnetic traps (Do and Ahn 2008). Active mixing by magnetic manipulation (Herrmann et al. 2006) or alternating flow directions (Diercks et al. 2009) can be added to passive trapping designs to accelerate binding.
The inclusion of an active retention system adds multiple sample renewability to these assays since the micro-beads can be flushed out and replaced for each new sample. This can be accomplished through pneumatic valving of channels (Yun et al. 2006; Yoo et al. 2011), external flow control (Iwai and Wei-Heong 2011), magnetic trapping with external valving (Choi et al. 2002), optical trapping (Tanaka et al. 2011), and other methods. Active retention devices allow for the possibility of assay automation when supported by a logic control system.
The design presented here differs from most renewable microfluidic bead assays reported in the literature in that the microbeads are incubated in a continuous flow environment, as opposed to being held static during incubation. The continuous flow operation is enabled by the use of paramagnetic microbeads as a functionalized assay surface. However, the assay automation does not require external logic, since the logic is instead inherent to the microfluidic design. In this alternate microbead incubation method, the microbeads are constantly propelled through the device by the background flow. The microbead trajectory is partially decoupled from the background flow using a magnetic field in order to transfer the beads between adjacent reagent streams. The magnetic force acting on the bead can be approximated as where B is the magnetic field from the magnet, μo is the magnetic permeability of free space, χ is the magnetic susceptibility of the bead (χ = 0.170) (Shevkoplyas et al. 2007), and Rc is the radius of the bead magnetic core (~0.5 μm). As described previously, the force on the microbead imparted by the magnetic field is approximately 7.5 pN while the shear force on the bead due to the background flow is greater than 22.3 pN so the beads are not statically trapped and can transverse through the device (Sasso et al. 2010). With proper tuning the device has been shown to achieve 100 % bead stream transfer efficiency.
The initial work in this field used magnetic microbeads which were pulled across a channel with stratified reagent streams patterned as adjacent colaminar flows, and an external permanent magnet which directed the microbeads across the channel (Peyman et al. 2008, 2009). This design enabled fully automated microbead incubation based entirely on passive control. With this approach, the force exerted on a paramagnetic microbead is proportional to the square of the magnetic field intensity gradient. Because the magnetic field intensity is empirically modeled to drop off with the distance from the magnet approximately to the third power (Garcia and Bonen 1999), the field gradient increases significantly closer to the magnet surface. Thus, as the bead is attracted towards the magnet, the tangential bead velocity increases and the residence time in a fixed width reagent stream decreases. This magnetic field distribution also implies that there is a limit to the reagent stream widths, which ultimately limits incubation time and therefore the limit of detection for a specific antigen. If the streams are made too wide, when paramagnetic beads enter a device at a distance furthest away from the magnet, both the field intensity and field gradient are very small so the force on the bead is not sufficient to attract the beads towards the magnet to produce the transverse motion of the bead. Finally, precise positioning of the external magnet has a large impact on the bead velocity through the device, which can also affect incubation residence time and repeatability, and can increase fabrication complexity related to magnet placement.
Later work used an alternate magnetic actuation method where microbeads were pulled across coplanar streams by external magnets to transfer beads from one environment to another, but the bead movement was constrained by the microchannel walls. The beads were then propelled along the channel by the shear from the perfusion flow, decoupling the bead incubation time from the magnetic actuation (Sasso et al. 2010). This design allowed improved control of the incubation time in each reagent stream, as well as simplified device fabrication. However, none of these designs allowed sufficient bead incubation times for the high sensitivity assays required in a broad range of clinical biomarker detection applications.
A new microdevice has been designed to significantly increase the allowable incubation time in an automated magnetic microbead assay to enable high sensitivity. In this new design, subsequent to introduction of the microbeads, each reagent stream is diverted into long spiral channels, so that the bead incubation times are significantly greater than have been previously reported for continuous flow micro-fluidic bead assays. The device presented is compatible with popular off-the-shelf paramagnetic microbead immunoassay kits such as those based on the Luminex® xMAP technology. Using a three-layer design with a single incubation step processed in each layer, the device continuously incubates beads through all three required incubation stages of the assay. The reagent sets allow multiplexing to detect and quantify up to 50 analytes within a single sample. This device can detect analytes within their clinically relevant concentration range with incubation times at a minimum of 5 min per incubation stage (15 min total incubation).
This automated microfluidic bead assays enables two important benefits which are not available with other available technologies. First, it enables rapid turnaround for single sample assays, with multiplexed results of high specificity immunoassays available within 20 min or less of sample extraction. These benefits are useful for point-of-care diagnostics as well as various research applications. Second, when coupled with a continuous detection flow cytometry system, the microdevice will allow continuous monitoring of time-varying sample concentrations at high sampling rates. This will enable direct on-line monitoring of analyte concentrations in unprocessed sample streams such as blood or cerebrospinal fluid (CSF), a feature which is not offered by any immunoassay technology currently on the market.
2 Operating principle
It has previously been demonstrated that a continuous flow microfluidic structure could perform all required incubation steps of a two-stage antigen sandwich immunoassay using paramagnetic microbeads (Sasso and Zahn 2009; Sasso et al. 2010). In this arrangement, a monoclonal antibody with specificity to the antigen of interest is conjugated to the microbeads’ surface using a variety of conjugation approaches prior to the assay. In the first stage of the assay, the microbeads are incubated with the sample, and they subsequently bind antigen with an amount proportional to the sample concentration. In the second stage, a fluorescently tagged secondary antibody which also has specificity to the antigen of interest is incubated with the beads. Following the incubation steps, the fluorescence intensity of the microbeads is correlated to the sample concentration through a calibration curve. The microdevice presented here adds a third incubation stage to accommodate assays where the fluorescent tag is added subsequent to the secondary antibody in a separate binding step, along with significantly longer incubation times than any continuous flow device previously reported in the literature.
2.1 Assay operation
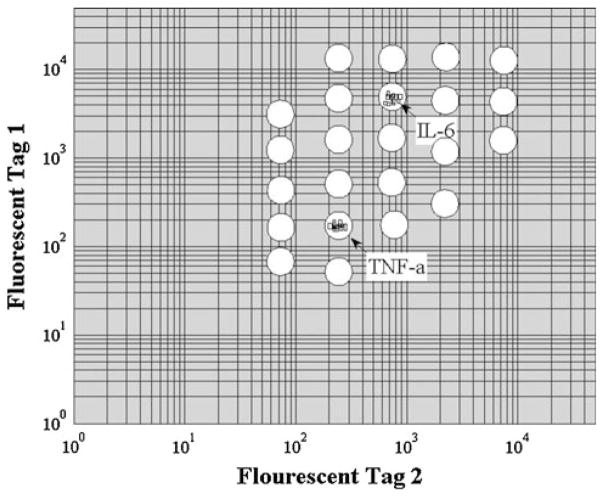
The Luminex® xMAP technology has been chosen in this work due to its multiplexing abilities and specificity for detecting a wide range of biomarkers; up to 50 simultaneous analytes can be detected in a single sample. Additionally, ‘blank’ carboxyl terminated beads are available for antibody conjugation through N-hydroxysulfosuccinimide (Sulfo-NHS) and 1-ethyl-3(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) chemistry for customization to link other biomarker specific antibodies to the beads. The magnetic Luminex® multiplex assays use optically encoded paramagnetic beads (6 μm in diameter) conjugated with biomarker antibodies. Each bead is encoded by a red and IR dye at varying intensity ratios for identification and gating in a two-color flow cytometry contour plot as illustrated in Fig. 1.
Fig. 1.
Representative fluorescent coding used in a Luminex® xMAP assay. Each circle represents a potential gating region for fluorescent detection of the analyte specifying colors. In this case, a duplex assay is shown, where fluorescent particle events are detected in the two regions of interest
The Luminex® assay uses a three-stage incubation approach. The first stage captures the antigen of interest by incubating the antibody coated microbeads with the sample, where the amount of bound antigen on each microbead after incubation is proportional to the sample concentration. In the second stage, the beads are incubated with a biotinylated secondary antibody to the antigen of interest to create the sandwich structure. Finally, in a third stage, the microbeads are fluorescently labeled by incubation with a streptavidin-phycoerythrin (PE) conjugate to fluorescently tag the detection antibody. The three-stage microbead assay is depicted in Fig. 2.
Fig. 2.
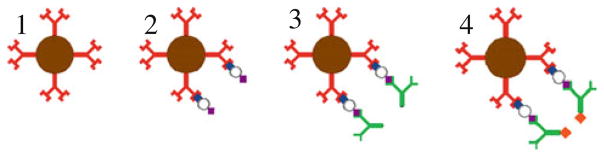
Schematic of microbead incubation. 1 Prior to assay, the magnetic microbead is conjugated with an antigen-specific antibody; 2 After the first incubation stage, the antigen is bound to the bead in proportion to its sample concentration; 3 A biotinylated secondary antibody binds to all antigen captured to each bead; 4 A streptavidin conjugated fluorophore binds to the secondary antibody
The fluorescence intensity of each bead after the three incubation stages is correlated to the antigen concentration in the sample through a calibration curve which is constructed from known antigen concentration standards. As with other immunoassay techniques, the calibration curve is constructed each time the assay is run to account for variations in binding efficiency, photobleaching of the fluorescent tag, and other factors which vary the bead fluorescence intensity at a given sample concentration. The sample concentration as well as the color-coded identifiers are detected via flow cytometry.
2.2 Microdevice operation
The microfluidic assay utilizes a magnetic separation scheme where microbeads are pulled from one reagent stream to the next by external magnets (Fig. 3). After stream transfer, the bead carrier solution flows into a waste outlet while the beads flow into the incubation spiral where the appropriate antigen/antibody binding can occur. A spiral incubation structure was used to allow a very long incubation channel while conserving device layout area. The incubation spiral is located away from the magnet so that the magnetic field does not have an effect on the beads while in the incubation channel, avoiding magnetic crosstalk between the beads and magnet after stream transfer. The total incubation time of the beads within the spiral can be adjusted by varying the perfusion flow rate. The flow rate must be chosen so that it is in a range where the fluid shear in the incubation channel is high enough to keep the beads moving without sticking to the channel walls. It also cannot flow so quickly that there is not enough time for the magnetic stream transfer to occur. Additionally, the separation channel should be designed such that its width creates an appropriate flow velocity in the range of volume flow rates expected to be used. The device as shown can be used at perfusion flow rates ranging between 200 nl/min and 2 μl/min, with a 100 μm wide transfer channel.
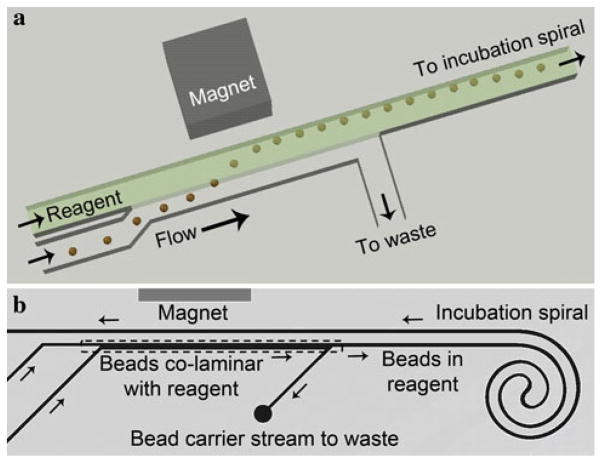
Fig. 3.
a Schematic of magnetically actuated bead transfer. The magnet pulls the beads from the carrier stream into the reagent stream, and the carrier stream is diverted to waste so that only the reagent stream with the microbeads continues to the incubation spiral. b Simplified layout of a single incubation device layer with bead transfer occurring within the layer. The bead carrier solution and incubation solution enter from the bottom left. The bead transfer area represented by the schematic above is within the dashed box. The carrier solution is then diverted to waste while the bead and reagent enters the incubation spiral. Following incubation, the beads pass by the magnet a second time where they can flow into a device outlet or transfer into another similar layer
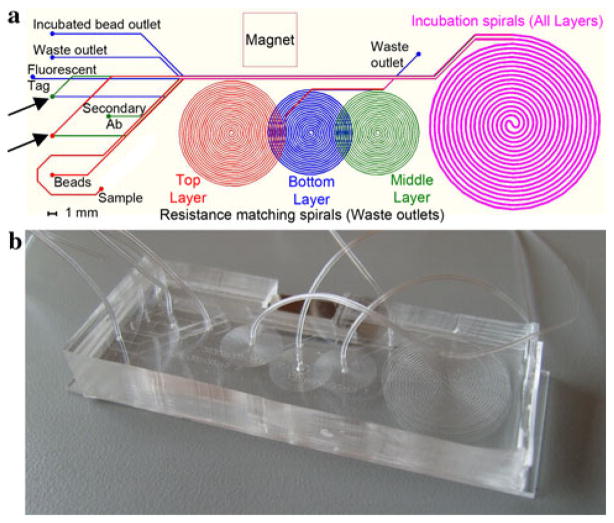
The device presented uses three layers with a single magnet to perform the entire three-stage assay (Fig. 4). The layers are aligned on top of each other so that the bead stream transfer occurs in the same region of the device in each layer, and a single magnet can be used for the entire device operation. The first layer has two inlets which create a laminar flow pattern between the initial bead carrier solution and the antigen sample in a common channel. The magnet pulls the beads across the flow boundary into the antigen stream. As the channel progresses, the initial bead carrier solution is diverted to waste while the beads continue in the antigen stream into the incubation spiral. A second spiral on the same layer balances the flow resistance to control the amount of fluid which is diverted to waste such that only the carrier solution is diverted to waste. Each resistance matching spiral is precisely designed to have the same flow resistance as the remainder of the device. Thus, on the top layer the matching spiral has the resistance of three incubation spirals, on the middle layer it has the resistance of two incubation spirals, and on the lower layer the matching spiral has equal resistance to a single incubation spiral. This arrangement diverts half of the total flow through the resistance matching channel to waste so that only the reagent stream enters the incubation spiral. Because the resistance of a channel depends on both its width and its length, the width of the resistance matching spiral is smaller than that of the incubation channel so that the total channel length can be reduced.
Fig. 4.
a Device layout, each of the smaller spirals performs flow resistance balancing on one of the three layers, and the large incubation spiral is identical on all layers. The top layer is shown in red, the middle layer in green, and the bottom layer in blue. Overlayed layers are shown in fuchsia. Layer transfers are depicted with arrows. b Photograph of complete three layer PDMS microdevice (color figure online)
The beads then transfer to the second layer where the process is repeated except now the antigen solution is replaced with the secondary antibody solution. Finally, the incubation is repeated once again in the third layer with the streptavidin-PE fluorescent tag. Following incubation beads are collected from the device and analyzed via flow cytometry. Thus, each data point represents the ensemble average fluorescence intensity of the beads in the sample incubated within the device.
It has been noted that following the stream splitting, beads tend to migrate away from the spiral wall. This is thought to be due to a slight flow imbalance caused as the carrier solution is diverted to waste. At each stage, after stream transfer, the beads have been found to travel within the incubation spiral at approximately the same velocity so each bead has approximately the same incubation residence time. Even though the flow profile is expected to be parabolic in the depthwise direction, the magnetic beads settle within the channel because they are higher density (ρbead ~ 2 g/ml) than the supporting fluid so each bead traverses the spiral at about the same channel depth. Across the channel, the flow velocity at this depth is approximately the same except close to the walls because the channel has an aspect ratio of 5:1 width-to-depth. Thus, since the each bead traverses the spiral at approximately the same velocity all beads have approximately the same incubation time.
3 Materials and methods
3.1 Microdevice fabrication
The chip was fabricated by standard soft lithography of PDMS (Duffy et al. 1998). The three layers are cast separately on SU-8 photoresist patterned substrates with a 40 μm channel height. The microchannel widths range from 50 to 200 μm, with the bead transfer region having a width of 100 μm, and the incubation spiral having a width and length of 200 μm and 60 cm, respectively. The tubing connection ports and layer transfer holes are punched through each layer with a sharpened 19 gauge needle resulting in ~1 mm holes. Layer alignment is done by eye, where only the layer transfer hole requires precise alignment accuracy for proper device operation, and there is a ±0.25 mm tolerance on this alignment. The top layer is first bonded to the middle layer using corona discharge activation followed by heating to 100 °C for 1 h. The middle layer ports are punched through both layers to allow insertion of tubing on the top of the chip surface. The lower layer is then bonded to the top-middle complex and its ports are punched through all three layers. Finally, a 75 mm ×25 mm glass microscope slide is bonded to the bottom of the lower layer. A location for the magnet is cut out with a razor blade such that the magnet is pressed into the material for retention. Tubing is pressed into the punched holes, which provides a sealed connection. The device layout and a photograph of a complete micro-device are shown in Fig. 4.
3.2 Experimental procedures
All experiments were based on the same assay reagent kit. A Bio-Plex Pro Magnetic Cytokine Assay (Bio-Rad, Hercules, CA) duplex based on Luminex® technology was selected with specificity to the cytokines TNF-α and IL-6, which have clinical significance in the inflammation cascade.
3.2.1 Reagent preparation
All reagents were included in the immunoassay kits and were used as instructed by the manufacturer with slight modifications. The reagents for the on-chip assay are prepared somewhat differently from the assay instructions to optimize them for microdevice infusion. 100 μl of each microbead stock is mixed in a microcentrifuge tube, washed twice with Wash Buffer, and suspended in 600 μl of Ficoll-Paque Plus (GE Healthcare, Wakesha, WI) for density matching such that the microbeads do not sediment after vortexing. 75 μl of each secondary detection antibody stock is mixed with 600 μl of antibody diluent. 6 μl of the streptavidin-phycoerythrin fluorescent tag stock is mixed with 600 μl of Assay Buffer. The provided wash buffer is used for the final microbead wash stream. All antigen samples were created by serial dilution of the provided Bio-Plex standards, and are in the general range 0.1–1,000 pg/ml. A 10× serial dilution was used for all on-chip assays as well as the bench top comparison data, while a 4× serial dilution was used for bench top experiments varying incubation time tests.
3.2.2 Fluorescence detection
For all experiments, the incubated bead samples were plated in standard 96-well plates, and interrogated using a Bio-Plex 200 flow cytometer. The machine uses two lasers (green 532 nm Nd-YAG, and red 635 nm laser diode) for detection of the three fluorescence channels (the two bead optical coding intensities and the bead PE labeling intensity) and one side scatter channel for doublet discrimination. All detectors use 15-bit analog to digital conversion. The doublet discriminator window was set at 8,000–24,000. Each sample infusion was 50 μl, and a minimum of 50 beads per analyte region was required (50 for TNF-α plus 50 for IL-6). Following analysis, the Bio-Plex 200 reports the mean bead fluorescence intensity but does not report the fluorescence variance. All experiments used the high sensitivity PMT mode except the detection limit test, which used the low sensitivity mode to achieve a wide dynamic range.
3.2.3 Off-chip assay detection limit test procedure
The assay was tested by varying the incubation times using an otherwise standard incubation procedure to characterize the binding kinetics and the resulting relationship between assay sensitivity and incubation time. The procedure from the assay manufacturer’s instructions (Bio-Plex Pro Assay Instruction Manual, Cytokine, Chemokine, and Growth Factors) was followed exactly, with the filter plate washing method, and using the provided assay standards. The incubation times were adjusted by delaying the addition of reagents to wells. For instance, in the first incubation stage, the standards were added at 0, 15, 20, 25, 28, and 29 min. The wash step then proceeded simultaneously for the entire plate. This procedure was followed for all three incubation stages. This procedure resulted in total incubation times of 70 min (30, 30, 10 min for stages 1, 2, and 3, respectively), 35 min (15, 15, 5), 23 min (10, 10, 3), 12 min (5, 5, 2), 5 min (2, 2, 1), and 3 min (1, 1, 1). The 70-min total incubation time is the incubation sequence recommended by the Bio-Rad instructions.
3.2.4 Off-chip benchmark assay procedure
In order to provide a benchmark to assess the performance of the microdevice, an analogous procedure to the on-chip assay was performed off-chip on the laboratory bench top. 100 μl of each microbead stock is mixed in a microcentrifuge tube, which serves as the incubation vessel, washed twice with Wash Buffer, aspirated and left unsuspended. 400 μl of antibody diluents is added to 50 μl of each detection antibody in a microcentrifuge tube. In another tube, 500 μl of assay buffer is mixed with 5 μl of streptavidin-PE stock solution. 50 μl of each standard is added to each incubation tube and the solution is mixed with a pipette. After 5 min, the incubation tubes are diluted with wash buffer and washed twice via centrifugation and left unsuspended. For the second incubation stage, 50 μl of the prepared detection antibody solution is added to each tube, mixed, and incubated for 5 minutes. The same wash procedure is used and finally 50 μl of the prepared streptavidin-PE fluorescent tag solution is added to the tubes and incubated for 5 min. After two more wash steps, the microbeads are suspended in 125 μl of assay buffer, vortexed, and plated for flow cytometric analysis.
3.2.5 On-chip assay procedure
The device is first infused via syringe pump with a mixture of PBS with 0.01 % (w/v) Bovine Serum Albumin (BSA) until all channels are filled. The reagents are then infused using two multi-infusion syringe pumps (PicoPlus 22 and PHD 2000, Harvard Apparatus, Holliston, MA), one pump for infusion of the magnetic microbeads, detection antibody, fluorescent tag, and wash buffer, and a second pump for the antigen sample syringe which is replaced repeatedly throughout the experiment to vary the sample concentration. All flow rates were set to 1 μl/min and beads were collected in fractions at the device outlet. This flow rate was found to result in a good balance between reagent utilization rate and fluid shear, since low shear conditions may allow beads to become stuck in the microchannels. Based on the cross sectional area of the incubation channel as described above, and the total channel length of 60 cm, the calculated average velocity results in an average incubation residence time of slightly <5 min. All bead collection is done in 1.5 ml microcentrifuge tubes. The collected beads are diluted with 100 μl of Assay Buffer and transferred to a well plate for flow cytometry fluorescence quantification.
Two experiments were conducted using this device: (1) generation of a calibration curve of fluorescence intensity as a function of sample concentration for comparison with beads incubated off chip on the bench top and (2) Temporal tracking of bead fluorescence to a time varying concentration input. When infusing a new sample concentration using the on-chip assay to generate a calibration curve, the older solution retained in the device was flushed prior to new sample collection. A pre-collection period was used in which each new sample is infused while the incubated bead outlet was not collected as a fraction for analysis. This ensures that the new sample is flushed completely through the system prior to bead collection. In this case, after a new sample syringe is attached, there is a 25-min pre-collection time, followed by 15 min of bead collection. For the second type of on-chip experiments where temporal data was acquired, there no pre-collection time was used, and the incubated beads were simply collected in 15-min fractions.
4 Results and discussion
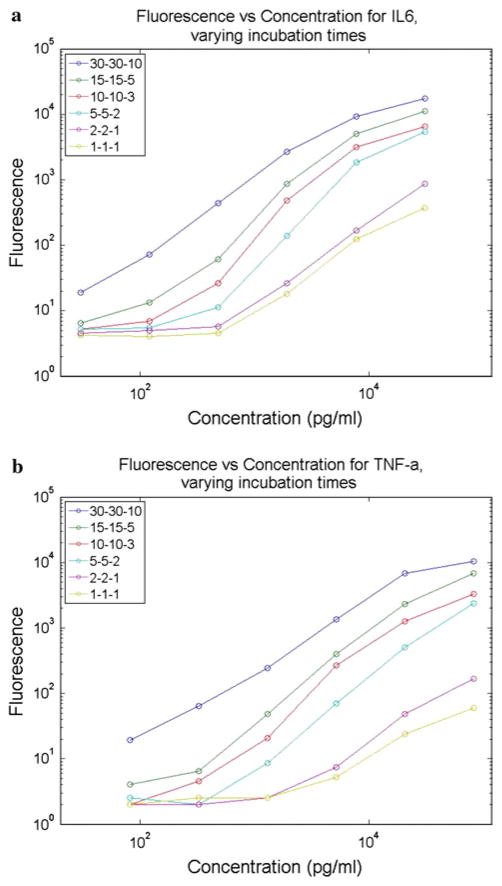
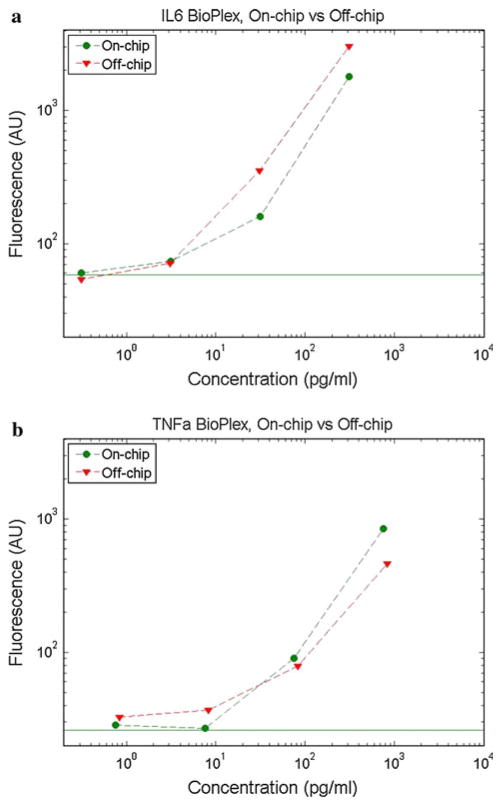
Initially, an empirical study was conducted to determine the effect of bead incubation time on bead fluorescence in the assay range 1 pg/ml to 100 ng/ml. By shortening incubation times, a faster assay turnaround time can be achieved at the expense of fluorescence intensity and/or assay sensitivity. As shown in Fig. 5, the mean fluorescence intensity of the microbeads increases with both the sample analyte concentration and the incubation time. This was tested using bench top incubations of the Bio-Plex assay with the provided standards, changing only the incubation times from the recommended procedure. Since a shorter total assay time is desirable for fast assay turnaround as well as for real-time monitoring, a 5-min per stage incubation time was chosen as a compromise between incubation time and assay sensitivity. This time was then used for both bench top (off-chip) and on-chip assays to compare their performance. The on-chip incubation device was specifically designed to allow a 5-min incubation time at a 1 μl/min flow rate. As shown in Fig. 6, the on-chip and off-chip assays yield similar bead fluorescence intensities at a given antigen concentration. In both cases, the two analytes were quantified simultaneously using the assay’s multiplexing feature. This data demonstrates that the microfluidic system can be applied as a direct automation platform for processing multiplexed microbead assays. A distinct calibration curve must be created each time a new assay is performed to account for expected interassay variations in bead fluorescence intensity due to differences in bead and labeling chemistry batches, binding kinetics, and fluorophore lifetime. The calibration curve is then used to correlate unknown samples to determine the sample concentration. The limits of detection are determined both by the position of the noise floor as well as the loss of slope in the bead fluorescence intensity curve at lower concentrations. The noise floor, as denoted by horizontal lines in Fig. 6, is found using a negative control where no analyte is present in the sample. The sample concentrations tested for IL-6 were 0.3, 3, 30, and 300 pg/ml, and for TNF-α were 0.8, 8, 80, and 800 pg/ml. Based on the plots, it is evident that while the noise floor is in the sub-pg/ml range, the data shows a very shallow slope in the single pg/ml range and thus the detection limit with this embodiment is around 10 pg/ml or slightly lower for IL-6 and 10s of pg/ml for TNF-α.
Fig. 5.
Results from bench-top testing of Bio-Plex assay for a IL-6 and b TNF-α with various incubation times. The three numbers represent the incubation time of each stage with 30-30-10 being the 70 min total incubation time recommended by the manufacturer
Fig. 6.
Representative multiplexed calibration curves comparing on-chip and off-chip Bio-Plex bead fluorescence intensity as a function of sample concentration for a IL-6 and b TNF-α multiplexed. The solid horizontal line represents the negative control level for the on-chip data. All incubation times were 5 min
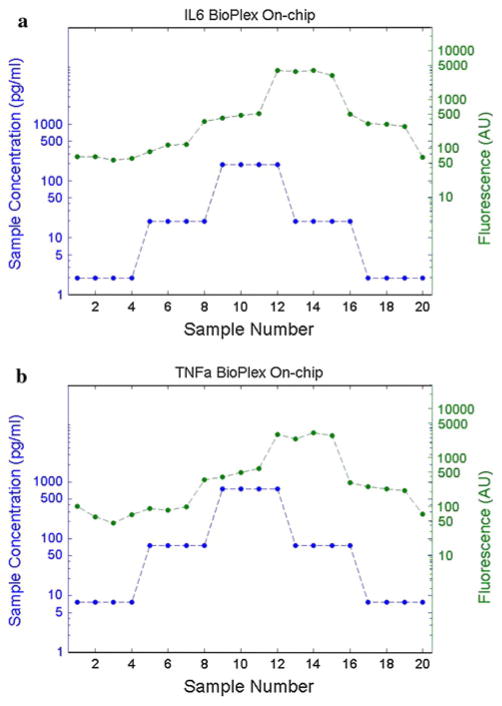
Additional experiments were performed to study the temporal response and repeatability of the microfluidic assay. These experiments used the same Bio-Plex assay for IL-6 and TNF-α as in previous experiments, but repeated sampling was used along with a step change in sample concentration. Figure 7 shows this temporal response data. The propagation delay, also referred to as the lag time of the assay, is evident in the shift between the inlet concentration trace and the measured fluorescence trace. Of particular concern is the ability of the assay to respond to both increasing and decreasing changes in concentration accurately and evenly. Variations in these measures could be caused by axial mixing of samples within the channel or by incomplete clearing along channel sidewalls causing a slowly recovering dilution of subsequent samples. The clearest test for these conditions is conducted by sampling identical sample concentrations in both the increasing and decreasing directions. The assay should provide an identical mean bead fluorescence intensity for the same sample concentrations regardless of the direction of change. It is expected that each change in sample concentration is followed by a time lag due to the propagation delay of incubation and dead volume within the outlet tubing. Based on the data in Fig. 7, the assay appears to respond appropriately without hysteresis.
Fig. 7.
Temporal response of microfluidic Bio-Plex assay for a IL-6 and b TNF-α multiplexed assay with step-wise changes in sample concentration. Incubated beads were collected every 15 min and analyzed using the Bio-Plex flow cytometer. The lower line represents the sample concentration at each time point, and the upper line represents the measured fluorescent intensity (AU). The expected assay propagation delay is evident
4.1 Incubation time and assay sensitivity
Given the complete three-stage incubation time of 15 min, the microfluidic assay could be applied to continuous monitoring applications where a lag time up to 20 min is acceptable, at a sample consumption rate of 1 μl/min. The dead volume within the layer transfer holes adds 5 min or less to the total lag time at this flow rate. This analysis delay still offers advantages over assays commonly used for clinical testing which can range from hours to even days for immunoassays, while providing not only short lag times but also very high sampling rates as beads and sample are continuously infused into the device. Furthermore, an integrated system could incorporate a cytometry flow cell on the chip, removing any analysis delay subsequent to incubation.
If higher sensitivity is needed, the incubation times can be lengthened to as much as 25 min per stage simply by reducing the infusion flow rate, at the expense of increased lag time, to provide detection limits below 1 pg/ml. For some applications, such as monitoring yield in bioproduction, the expected protein concentrations will be at least 100 times greater than typical clinical concentrations, and can be as much as 106 times greater for high yield processes. In these cases, the incubation times may be reduced to below 1 min per stage. If the incubation time must be reduced without increasing the sample flow rate, the spiral incubation channel can be shortened as necessary to reduce the total channel length.
4.2 Applications
This technology offers two major functionalities of interest for commercial applications. First, as an automation system, it can be applied in both clinical and laboratory settings. Clinical assays for disease biomarkers would benefit from the fully enclosed, hands-free operation which would reduce the possibility of sample handling errors, improve turnaround time, and remove the need for highly skilled technicians to perform assays. Research applications will also benefit from this type of automation especially when sequential sampling is involved. Additionally, this technology is a lower cost and lower maintenance alternative to existing robotic assay systems.
The second important feature of this technology is its ability to continuously monitor a sample with a time-varying analyte concentration. This feature sets this technology apart from almost all other assay automation technologies. An especially unique feature is that the assay is indefinitely renewable as long as there is a supply of fresh reagents to infuse, which allows for extended measurement times. The continuous operating mode is ideal for applications where sample concentrations are changing but only small volumes of the sample fluid are available for testing. This will enable monitoring of inflammation biomarkers in blood during surgeries where current sampling techniques require sample volumes which are too large to allow high sampling rates (Yang et al. 2007; Sasso et al. 2010; Aran et al. 2011). This function is also applicable to monitoring of bioprocesses such as biologics drug production.
4.3 Supporting instrumentation
For any of these applications, the microfluidic assay will require supporting instrumentation to control reagent and sample infusion and for fluorescence detection. The simplest embodiment would include a syringe based pumping system for discrete sampling, in which case a separate flow cytometer could be used for detection of the mean fluorescence intensity of incubated bead batches. Direct integration of a flow cytometer with the control system and chip would further enhance usability and decrease assay turnaround time, as well as enabling continuous monitoring applications. A well plate sipper system, often used on auto-sampling flow cytometry equipment, can be incorporated for serial processing of discrete samples. Finally, real time monitoring will require a flow control system for steady infusion of the sample into the microfluidic device. A moving average of bead intensities will produce a near real-time signal. With advances in laser diode and photo-detector technologies, it may eventually be possible to produce an easily portable or even hand-held version of this system.
5 Conclusions
This work has demonstrated a robust laboratory automation platform using microfluidics to execute an important microbead assay. The assay works with existing off-the-shelf reagents for a large variety of clinical and life science testing applications and customizable reagents are currently available which allow detection of many biomarkers in parallel. This highly versatile technology enables automated assaying with multiplexing and high sensitivity in all of these applications without the need for large robotic systems or complex external actuators. It is especially well suited for applications where a small number of samples must be assayed with rapid results, and where continuous concentration monitoring is required. Future work will involve combining the microfluidic platform with a flow cytometry system to include automation of the detection step and to demonstrate continuous monitoring.
Acknowledgments
This work was funded by grants from the New Jersey Commission on Spinal Cord Research, the National Institutes of Health Biotechnology Pre-doctoral Training Program, the National Heart Lung Blood Institute, the Wallace H. Coulter Foundation Early Career Translational Research Awards in Biomedical Engineering, and the NIH funded Point-of-Care Center for Emerging Neuro-Technologies.
Contributor Information
Lawrence A. Sasso, Email: Lsasso@eden.rutgers.edu, BioMEMS Laboratory, Department of Biomedical Engineering, Rutgers, The State University of New Jersey, 599 Taylor Road, Piscataway, NJ 08854, USA
Ian H. Johnston, BioMEMS Laboratory, Department of Biomedical Engineering, Rutgers, The State University of New Jersey, 599 Taylor Road, Piscataway, NJ 08854, USA
Mingde Zheng, BioMEMS Laboratory, Department of Biomedical Engineering, Rutgers, The State University of New Jersey, 599 Taylor Road, Piscataway, NJ 08854, USA.
Rohit K. Gupte, BioMEMS Laboratory, Department of Biomedical Engineering, Rutgers, The State University of New Jersey, 599 Taylor Road, Piscataway, NJ 08854, USA
Akif Ündar, Email: aundar@psu.edu, Penn State Hershey Pediatric Cardiovascular Research Center, Department of Pediatrics, Surgery, and Bioengineering, Penn State Milton S. Hershey Medical Center, Penn State College of Medicine, Penn State Children’s Hospital, Hershey, PA, 17033-0850, USA.
Jeffrey D. Zahn, Email: jdzahn@rci.rutgers.edu, Department of Biomedical Engineering, Rutgers, The State University of New Jersey, Room 311, 599 Taylor Road, Piscataway, NJ 08854, USA
References
- Andersson H, van der Wijngaart W, et al. Micromachined flow-through filter-chamber for chemical reactions on beads. Sens Actuators B Chem. 2000;67(1–2):203–208. [Google Scholar]
- Aran K, Fok A, et al. Microfiltration platform for continuous blood plasma protein extraction from whole blood during cardiac surgery. Lab Chip. 2011;11(17):2858–2868. doi: 10.1039/c1lc20080a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breadmore MC, Wolfe KA, et al. Microchip-based purification of DNA from biological samples. Anal Chem. 2003;75(8):1880–1886. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- Choi JW, Oh KW, et al. An integrated microfluidic biochemical detection system for protein analysis with magnetic bead-based sampling capabilities. Lab Chip. 2002;2(1):27–30. doi: 10.1039/b107540n. [DOI] [PubMed] [Google Scholar]
- Diercks AH, Ozinsky A, et al. A microfluidic device for multiplexed protein detection in nano-liter volumes. Anal Biochem. 2009;386(1):30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do J, Ahn CH. A polymer lab-on-a-chip for magnetic immunoassay with on-chip sampling and detection capabilities. Lab Chip. 2008;8(4):542–549. doi: 10.1039/b715569g. [DOI] [PubMed] [Google Scholar]
- Duffy DC, McDonald JC, et al. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Garcia AA, Bonen M. Bioseparation process science. Wiley-Blackwell; UK: 1999. [Google Scholar]
- Herrmann M, Veres T, et al. Enzymatically-generated fluorescent detection in micro-channels with internal magnetic mixing for the development of parallel microfluidic ELISA. Lab Chip. 2006;6(4):555–560. doi: 10.1039/b516031f. [DOI] [PubMed] [Google Scholar]
- Iwai K, Wei-Heong T, et al. A resettable dynamic microarray device. Biomed Microdev. 2011;13:1089–1094. doi: 10.1007/s10544-011-9578-7. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Choi K, et al. PDMS micro bead cage reactor for the detection of alpha feto protein (AFP) Sens Actuators B Chem. 2008;128(2):349–358. [Google Scholar]
- Oleschuk RD, Shultz-Lockyear LL, et al. Trapping of bead-based reagents within microfluidic systems: on-chip solid-phase extraction and electrochromatography. Anal Chem. 2000;72(3):585–590. doi: 10.1021/ac990751n. [DOI] [PubMed] [Google Scholar]
- Peyman SA, Iles A, et al. Rapid on-chip multi-step (bio)chemical procedures in continuous flow—manoeuvring particles through co-laminar reagent streams. Chem Commun. 2008;14(10):1220–1222. doi: 10.1039/b716532c. [DOI] [PubMed] [Google Scholar]
- Peyman SA, Iles A, et al. Mobile magnetic particles as solid-supports for rapid surface-based bioanalysis in continuous flow. Lab Chip. 2009;9(21):3110–3117. doi: 10.1039/b904724g. [DOI] [PubMed] [Google Scholar]
- Sasso LA, Undar A, et al. Autonomous magnetically actuated continuous flow microimmunofluorocytometry assay. Microfluid Nanofluid. 2010;9(2):253–265. doi: 10.1007/s10404-009-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso LA, Zahn JD. The thirteenth international conference on miniaturized systems for chemistry and life sciences; Jeju, Korea. 2009. [Google Scholar]
- Sato K, Tokeshi M, et al. Integration of an immunosorbent assay system: analysis of secretory human immunoglobulin A on polystyrene beads in a microchip. Anal Chem. 2000;72(6):1144–1147. doi: 10.1021/ac991151r. [DOI] [PubMed] [Google Scholar]
- Sato K, Yamanaka M, et al. Microchip-based enzyme-linked immunosorbent assay (microELISA) system with thermal lens detection. Lab Chip. 2004;4(6):570–575. doi: 10.1039/b411356j. [DOI] [PubMed] [Google Scholar]
- Shevkoplyas SS, Siegel AC, et al. The force acting on a superparamagnetic bead due to an applied magnetic field. Lab Chip. 2007;7(10):1294–1302. doi: 10.1039/b705045c. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tsutsui S, et al. Hybrid optical tweezers for dynamic micro-bead arrays. Opt Express. 2011;19(16):15445–15451. doi: 10.1364/OE.19.015445. [DOI] [PubMed] [Google Scholar]
- Thompson JA, Du XG, et al. Polymeric microbead arrays for microfluidic applications. J Micromechan Microeng. 2010;20(11) [Google Scholar]
- Yang S, Undar A, et al. Continuous cytometric bead processing within a microfluidic device for bead based sensing platforms. Lab Chip. 2007;7:588–595. doi: 10.1039/b703808a. [DOI] [PubMed] [Google Scholar]
- Yoo SK, Kim YM, et al. Bead packing and release using flexible polydimethylsiloxane membrane for semi-continuous biosensing. Artif Organs. 2011;35(7):E136–E144. doi: 10.1111/j.1525-1594.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- Yun KS, Lee D, et al. A microfluidic chip for measurement of biomolecules using a microbead-based quantum dot fluorescence assay. Measur Sci Technol. 2006;17(12):3178–3183. [Google Scholar]