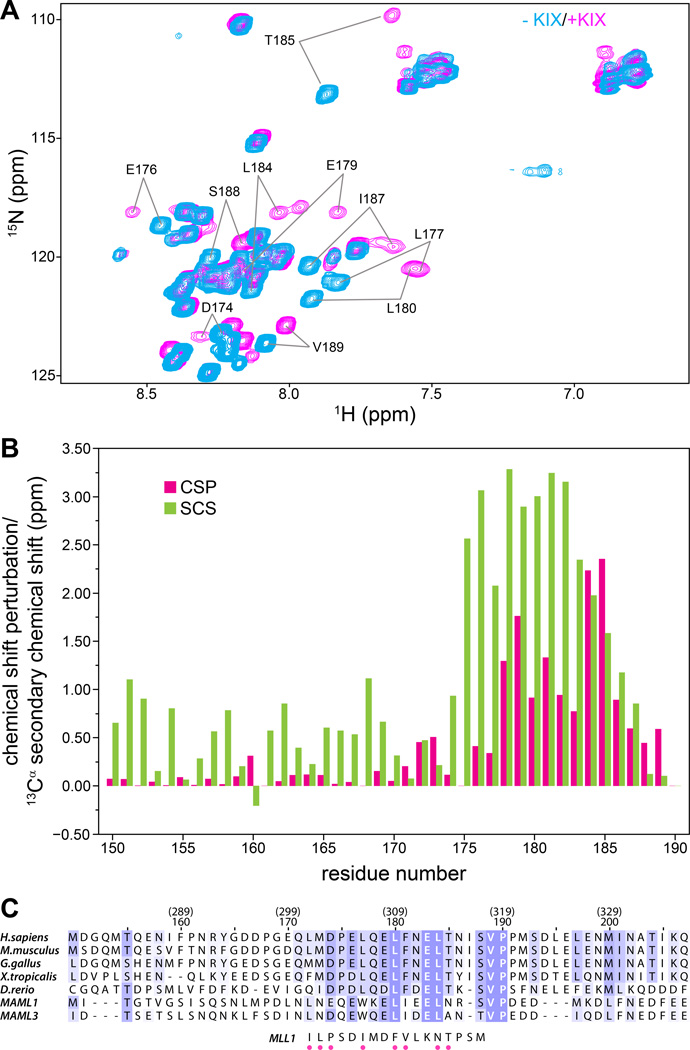
Figure 2. The CRTC1-MAML2 TAD1 binds KIX via a short conserved, helical segment.
(A) 1H-15N correlated spectra of 15N,13C-labeled CRCT1-MAML2 TAD150–190 recorded in the absence (cyan) and presence (magenta) of one equivalent of CBP KIX at 25 °C. (B) Backbone amide CSPs (green) for CRTC1-MAML2 TAD150–190 induced by the addition of one equivalent of KIX plotted as a function of residue number; the TAD concentration was 0.7 mM. CSPs were calculated as described in Figure 1B. Also graphed are 13Cα secondary chemical shifts (SCS; red) for TAD150–190 when bound to KIX as a function of residue number. (C) CLUSTAL Ω-guided multiple sequence alignment of MAML2 homologues from various species and comparison with the sequence of the MLL1 segment that engages KIX. The residue numbering in the context of human MAML2 is given in parenthesis on top of the numbering in the context of the CRTC1-MAML2 fusion. Sequences were shaded based on substitution scores from the BLOSUM62 scoring matrix in JalView.(31) MLL1 residues that make direct contacts with KIX in the NMR structure(25) are identified by dots below the sequence.