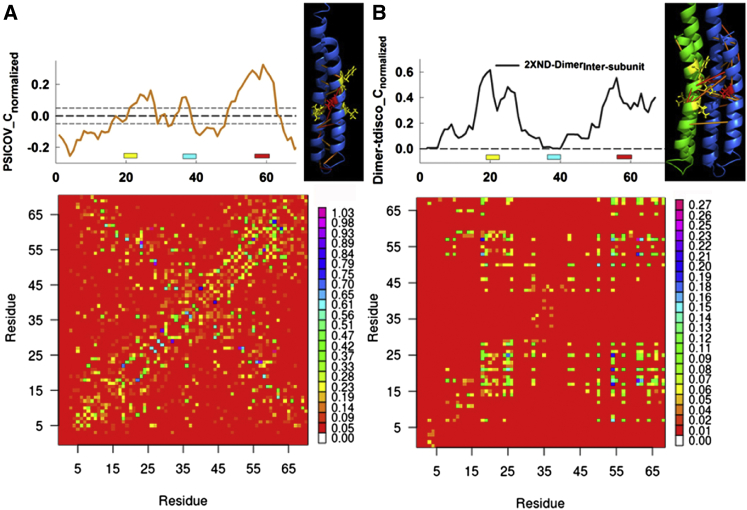
Figure 6.
Relation between subunit architecture and residue coevolution. (A) The PSICOV coevolution matrix for the trimmed MSA, based on the PDB: 2XND subunit sequence. Matrix elements are color-coded based on correlation strength normalized between 0 and 1. The matrix centrality profile, corrected for residual correlations, is on top. (Short dashed lines) Either side of the zero mean (dashed line) difference shows deviation (±1σ) scaled to the variation in the mean randomized library profile (Fig. S4). The PDB: 2XND subunit map adjacent to the centrality profile shows the top-scoring correlations (blue, weak to red, strong) cluster between the essential E57 glutamate (red) and IB-site residue I24, L55, M59, and F62 (yellow) side chains. (B) The intersubunit, dynamic correlation matrix for the isolated PDB: 2XND dimer (see Fig. S4 for full matrix). Matrix elements are color-coded according to their nMI value. The PDB: 2XND dimer, with color-coded subunits, adjacent to the centrality profile shows the strongest correlations (nMI > 0.20), color-coded as in (A). The E57 and IB-site residues are now in proximity. (Yellow and red bars) GGGG-motif and IB-site positions as in Fig. 3. (Cyan bar) Position of the conserved cytoplasmic loop. To see this figure in color, go online.