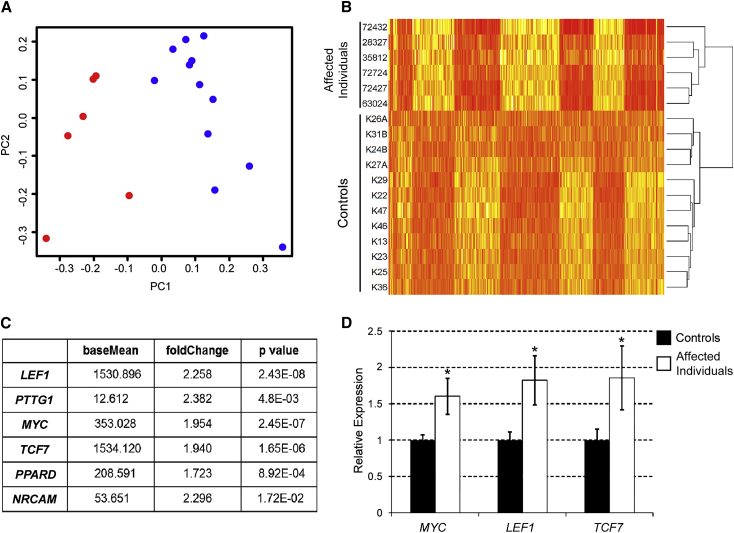
Figure 1.
Differential Gene Expression between ID-Affected Individuals and Control Groups
(A) Principal component analysis of RNA-Seq data showing the first two principal components (PC1 and PC2) of six ID-affected individuals (red) and twelve control individuals (blue) from RPKM values of 24.364 genes.
(B) Heat map showing differential expression patterns between ID-affected and control individuals with 452 genes with a fold change >2 and p value < 0.01 used in pathway analysis. Higher relative expression is indicated by darker hues.
(C) Mean normalized expression values (baseMean), fold change, and negative binomial p value for six Wnt/β-catenin target genes with significant upregulation in ID-affected individuals in comparison to control individuals.
(D) Validation of RNA-Seq data with qRT-PCR in control and ID-affected individuals for three Wnt/β-catenin target genes: MYC, LEF1, and TCF7. Relative expression shows the average values of twelve control individuals (set at a value of 1) and six ID-affected individuals. Error bars represent SEM. Asterisks denote a p value of <0.05.