Abstract
Numerous studies focusing on genetic variants in order to find cetuximab subpopulation biomarkers have emerged, yet the significance of each biomarker is diverse. Based on these results, we carried out a meta-analysis to assess the correlation between epidermal growth factor (EGF) A61G polymorphism and clinical outcomes of metastatic colorectal cancer (mCRC) patients treated with cetuximab. We aim to prove that EGF polymorphisms may be potential biomarkers for cetuximab therapeutic strategies. We identified 6 previously published studies including 569 patients treated with cetuximab-based regimens. Outcomes included clinical response, progression-free survival (PFS), and overall survival (OS). GG homozygote showed association with better response rates (GG vs. AA+AG, OR = 2.82; 95% CI = 1.58-5.04) and than AA+AG genotypes. This meta-analysis showed that mCRC patients harboring GG genotype of EGF A61G polymorphism inclined to have a better response rate with cetuximab treatment.
Keywords: EGF, polymorphism, mCRC
Introduction
Recently, the development of novel therapeutic agents that specifically target growth factor pathways has triggered great interest. Such targeted agents may offer alternative treatments for patients sensitive or refractory to standard chemotherapy. In particular, agents targeting members of the epidermal growth factor receptor (EGFR) have shown promising therapeutic efficacy [1].
Nowadays, two different anti-EGFR targeting regimens are clinically available: small molecule inhibitors in intracellular phosphotyrosine kinase domain; and monoclonal antibodies targeting the extracellular domain of the receptor. As the most extensively investigated anti-EGFR monoclonal antibody, cetuximab is currently approved as a single agent or in combination with irinotecan for the treatment of patients with metastatic colorectal cancer (mCRC). Cetuximab is a chimeric monoclonal G1 (IgG1) antibody that sticks to the EGFR with high affinity. The antibody down-regulates surface EGFR expression through blocking ligand binding and inducing receptor internalization and degradation [2].
Although cetuximab has shown effectiveness in metastatic colorectal cancer and has been approved for use in combination with irinotecan in chemorefractory patients [3,4], clinical assessments showed that the response rate was only 30% in patients with mCRC [5]. In view of this low response rate, investigators were spurred on into identifying mechanisms that can improve the response rate, if only in a subgroup of patients with mCRC. As known to all, EGFR expressed in colorectal cancer has been regarded as a poor prognostic factor correlating with aggressive disease and decreased survival [6,7]. Therefore upon first entering the clinic, cetuximab trials were conducted in patients whose tumors were tested positive for EGFR expression by immunohistochemistry. However, studies failed to demonstrate a consistent relationship between EGFR expression and response rate to cetuximab therapy [5,8]. With these findings, the National Comprehensive Cancer Network shut the door on the use of EGFR-expression test as criteria for selecting patients for cetuximab therapy [3,9].
Gene variants have been associated with response to cetuximab in mCRC patients, including gene copy number of EGFR [10,11]; and single nucleotide polymorphism (SNP) on the encode gene of EGF. Functional variant in the EGF 5’-untranslated region (EGF A61G, rs4444903) [12] has been regarded as modulating the EGFR ligand EGF, resulting in correlation with sensitivity to EGFR-targeted therapy. But the true relationship between them remains an important focus of evidence-based investigations.
Herein, we present the findings of a meta-analysis, attempting to assess the association between EGF A61G polymorphism and clinical outcomes of mCRC patients treated with cetuximab. We aim to show that EGF polymorphisms may be the potential biomarkers for cetuximab therapeutic strategies.
Materials and methods
This study was conducted in accordance with the ‘preferred reporting items for systematic reviews and meta-analyses’ (PRISMA) guidelines.
Study eligibility and identification
We did systematic computerized search in the PubMed and EMBASE databases, using following terms: ‘cetuximab’, ‘colorectal neoplasm’, in combination with ‘polymorphism’. The search details in PubMed database were as follows: (“cetuximab” [Supplementary Concept] OR “cetuximab” [All Fields]) AND (“colorectal neoplasms” [MeSH Terms] OR (“colorectal” [All Fields] AND “neoplasms” [All Fields]) OR “colorectal neoplasms” [All Fields] OR (“colorectal” [All Fields] AND “neoplasm” [All Fields]) OR “colorectal neoplasm” [All Fields]) AND (“polymorphism, genetic” [MeSH Terms] OR (“polymorphism” [All Fields] AND “genetic” [All Fields]) OR “genetic polymorphism” [All Fields] OR “polymorphism” [All Fields]). References in the retrieved articles were further screened for earlier original studies. The inclusion criteria were as followings: patients with histopathologically confirmed metastatic colorectal cancer, cetuximab-based therapies, comparisons of clinical response ratio among different genotypes of EGF A61G polymorphism, and relationship between this polymorphism and prognosis. When datasets were incomplete for necessary data, the corresponding authors were contacted to obtain missing information. Some studies were excluded if critical information was still missing after requests.
Data extraction and quality assessment
The following information was recorded from each recovered article: first author, journal and year of publication, number of patients analyzed, method of genotype discrimination, previous treatment, strategies of cetuximab administration and data linking variants to treatment outcomes.
We assessed the study quality in a descriptive and qualitative approach rather than a quantitative one, with regard to the following aspects, which were largely consistent with REMARK (Reporting recommendations for tumor marker prognostic studies) guidelines [13].
Data extraction and quality assessment of each study were conducted by two authors (Xiaoli Lu and Xiaowan Chen) independently, and discrepancies were resolved by consensus including a third author (Jingxu Sun).
Statistical analysis
The primary end point was clinical response rate, which was defined as the rate of complete response and the partial response rate. The relationship between EGF polymorphism and clinical response rate was assessed by odds ratio (OR) and corresponding 95% confidence intervals (CIs). A total of six genetic models, with three main models (M1, allele comparison, A vs. a; M2, recessive model, AA vs. Aa+aa; or M3, dominant model, AA+Aa vs. aa) and three models of multiple pairwise comparisons (M4, AA vs. aa; M5, Aa vs. aa; or M6, AA vs. Aa) were considered in this meta-analysis. Models M1 to M3 were considered as the primary genetic models of interest [14]. The risk ratio (RR) with 95% CI was used to assess the association between these polymorphisms and cetuximab-related skin toxicity (grades 0-1 versus 2-3).
The secondary end points were progression-free survival (PFS) and overall survival (OS). Hazard ratios (HRs) and their 95% CIs were combined to give an effective value for the quantitative aggregation of survival results. They were estimated from available data using the methods reported by Tierney [15], if these statistical variables were not given explicitly in an article.
Between-study heterogeneity was estimated using the χ2-based Q statistic [16]. Heterogeneity was considered statistically significant when p for heterogeneity < 0.05 and/or I2 > 50%. Fixed-effects model was used if heterogeneity did not exist; otherwise, a random-effects model was used. Subgroup-stratification analysis was appraised based on the therapeutic regimen of cetuximab, such as whether cetuximab was used as a single agent or in combination with the chemotherapy, and whether patients received cetuximab-based treatment as initial therapy or after refractory to chemotherapy. Finally, potential publication biases were evaluated in both Begg’s and Egger’s tests. A two-tailed p value of < 0.05 was considered statistically significant [17]. All data were analyzed using RevMan 5.2 analysis software (The Cochrane Collaboration) and STATA 12.0 analysis software (Stata Corporation, College Station, TX, USA).
Results
Summary of included and excluded studies
A total of 120 studies were initially identified (Figure 1). Among these, 48 were duplicates and 65 further studies were excluded because they did not include polymorphisms of interest; or did not assess any pharmacokinetic or prognostic outcomes of interest. Six studies met the inclusion criteria and were included in the final analysis [18-23]. The eligible studies and the corresponding characteristics are presented in Table 1.
Figure 1.
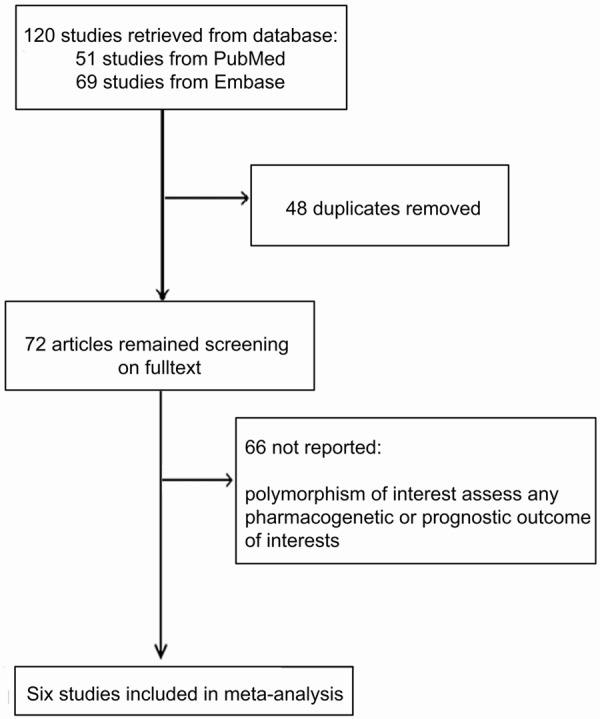
Flowchart of the studies selected.
Table 1.
Characteristics of eligible studies considered in the report
| Author_Year | Genetic variant | Patients included, n | Study design | Genotype analysis | Previous treatment | Study treatment | Response criteria |
|---|---|---|---|---|---|---|---|
| Zhang_2006 [22] (IMCL-0144) | EGF A61G | 34 | Retrospective† | PCR-RFLP | ≥ 2 chemotherapy | C alone | WHO |
| Graziano-2008 [19] | EGF A61G | 110 | Prospective | PCR-RFLP | ≥ 1 chemotherapy | C + I | RECIST |
| Lurje-2008 (IMCL-0144) [21] | EGF A61G | 116 | Retrospective† | PCR-RFLP | ≥ 2 chemotherapy | C alone | WHO |
| Garm-2009 [18] | EGF A61G | 71 | Retrospective | Taq-PCR | ≥ 1 chemotherapy | C + I | RECIST |
| Pander-2010 (CAIRO2) [23] | EGF A61G | 120 | Retrospective† | Taq-PCR | untreated | C + O | RECIST |
| Hu-2011 (multicenter) [20] | EGF A61G | 118 | Retrospective† | PCR-RFLP | RT | C + O | Dworak |
C alone: cetuximab used as single agent; C + I: cetuximab used in combination with irinotecan; C + O: cetuximab used in combination with oxaliplatin; RECIST: Response Evaluation Criteria in Solid Tumors; PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism.
EGF A61G polymorphism (rs4444903) and clinical response to cetuximab
Data concerning the predictive value of EGF A61G with respect to the sensitivity of mCRC to cetuximab-based treatment were available in 5 trials [18-21,23]. These covered 449 individuals. In allele comparison (M1), the 61G allele was more associated with better response rate than the 61A allele (G vs. A, OR = 2.11; 95% CI = 1.47-3.03; I2 = 46%). Moreover, in recessive model (M2), the GG homozygote showed association with a better response rate than AA+AG genotypes (GG vs. AA+AG, OR = 2.82; 95% CI = 1.58-5.04; I2 = 19%). While In dominant model (M3), no significant difference of response rate between AA homozygote and AG+GG genotypes were shown (GG+AG vs. AA, OR = 2.07; 95% CI = 0.74-5.79; I2 = 55%). In M4 model, GG homozygote showed significant association with a better response rate than the AA homozygote (GG vs. AA, OR = 3.91; 95% CI = 1.94-7.90; I2 = 41%). In M5 model, there was no significant difference between AA homozygote and AG heterozygote in terms of response rates (AG vs. AA, OR = 1.54; 95% CI = 0.49-4.91; I2 = 57%). However, in M6 model, GG homozygote showed significant association with better response rate than the AG heterozygote (GG vs. AG, OR = 2.25; 95% CI = 1.19-4.25; I2 = 39%). All pooled analysis findings were tabulated in Table 2.
Table 2.
Analysis of the association between EGF A61G and odds ratio of clinical response rate in main models
| M1: G vs. A | M2: GG vs. AG+AA | M3: GG+AG vs. AA | ||||||||
|
|
|
|
||||||||
| Study groups | Studies Numbers | OR (95% CI) | P | I2 | OR (95% CI) | P | I2 | OR (95% CI) | P | I2 |
|
| ||||||||||
| Overall | 5 | 2.11 (1.47, 3.03) | 0.00 | 46% | 2.82 (1.58, 5.04) | 0.00 | 19% | 2.07 (0.74, 5.79) | 0.16 | 55% |
| Therapy regimens | ||||||||||
| C alone | 2 | 1.69 (0.76, 3.74) | 0.20 | 67% | 2.87 (0.89, 9.23) | 0.08 | 0% | 0.72 (0.03, 16.99) | 0.84 | 70% |
| C combine | 3 | 2.24 (1.50, 3.36) | 0.00 | 55% | 2.81 (1.44, 5.48) | 0.00 | 57% | 2.75 (0.79, 9.55) | 0.11 | 64% |
|
| ||||||||||
| M4: GG vs AA | M5: AG vs. AA | M6: GG vs.AG | ||||||||
|
|
|
|
||||||||
| Study groups | Studies Numbers | HR (95% CI) | P | I2 | HR (95% CI) | P | I2 | HR (95% CI) | P | I2 |
|
| ||||||||||
| Overall | 5 | 3.91 (1.94, 7.90) | 0.00 | 41% | 1.54 (0.49, 4.91) | 0.46 | 57% | 2.25 (1.19, 4.25) | 0.01 | 39% |
| Therapy regimens | ||||||||||
| C alone | 2 | 2.68 (0.68, 10.55) | 0.16 | 31% | 0.68 (0.05, 8.39) | 0.76 | 52% | 2.62 (0.64, 10.77) | 0.18 | 0% |
| C combine | 3 | 4.49 (1.97, 10.23) | 0.00 | 60% | 2.17 (0.46, 10.23) | 0.33 | 71% | 2.16 (1.06, 4.41) | 0.03 | 59% |
C alone: cetuximab used as single agent; C combine: cetuximab used in combination with irinotecan or oxaliplatin based chemotherapy.
EGF A61G polymorphism (rs4444903) and PFS
A total of 4 studies targeted the relationship between EGF A61G polymorphism and PFS in dominant model (M3) [18-20,22]. Pooled analysis indicated that there was no significant association (GG+AG vs. AA, HR = 0.80; 95% CI = 0.54-1.18; I2 = 72%) between polymorphism and PFS in mCRC patients treated with cetuximab. In M4 model, no significant association between PFS and the EGF polymorphism homozygotes (GG vs. AA, HR = 0.65; 95% CI = 0.36-1.31; I2 = 63%). While in M5 model, a significant association was presented that AG heterozygote was related to better PFS than the AA homozygote (AG vs. AA, HR = 0.72; 95% CI = 0.54-0.96; I2 = 35%). All pooled analysis findings were tabulated in Table 3.
Table 3.
Analysis of the association between EGF A61G and progression-free survival (PFS) in main models
| M3: GG+AG vs. AA | M4: GG vs. AA | M5: AG vs. AA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Study groups | Studies Numbers | HR (95% CI) | P | I2 | HR (95% CI) | P | I2 | HR (95% CI) | P | I2 |
| Overall | 4 | 0.80 (0.54-1.18) | 0.26 | 72% | 0.65 (0.36-1.31) | 0.25 | 63% | 0.72 (0.54-0.96) | 0.02 | 35% |
| Therapy regimens | ||||||||||
| C alone | 2 | 0.86 (0.32-2.34) | 0.76 | 87% | 1.01 (0.27-3.78) | 0.99 | 76% | 0.83 (0.57-1.20) | 0.02 | 41% |
| C combine | 1 | 0.80 (0.58-1.20) | 0.28 | 61% | 0.47 (0.27-0.81) | 0.01 | -- | 0.58 (0.37-0.92) | 0.32 | -- |
C alone: cetuximab used as single agent; C combine: cetuximab used in combination with irinotecan or oxaliplatin based chemotherapy.
EGF A61G polymorphism (rs4444903) and OS
Data concerning the relationship between EGF A61G polymorphism and OS was investigated in 3 studies [18-20]. In dominant model (M3), the variant showed no association with OS (GG+AG vs. AA, HR = 0.99; 95% CI = 0.49-1.99; I2 = 86%). In M4 model, no significant relationship was found between genetic variant and OS (GG vs. AA, HR = 1.25; 95% CI = 0.85-1.84). In M5 model, negative association was shown (AG vs. GG, HR = 1.12; 95% CI = 0.89-1.41). In M6 model, there was also no significant association between polymorphism and better OS (AA vs. AG, HR = 1.09; 95% CI = 0.8-1.50). All pooled analysis findings were tabulated in Table 4.
Table 4.
Analysis of the association between EGF A61G and overall survival (OS) in main models
| M3: GG+AG vs. AA | M4: GG vs. AA | M5: AG vs. AA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Study groups | Studies Numbers | HR (95% CI) | P | I2 | HR (95% CI) | P | I2 | HR (95% CI) | P | I2 |
| Overall | 3 | 0.99 (0.49-1.99) | 0.97 | 86% | 0.98 (0.41-2.38) | 0.97 | 77% | 0.97 (0.53-1.77) | 0.91 | 70% |
| Therapy regimens | ||||||||||
| C alone | 2 | 1.37 (0.75-2.50) | 0.31 | 62% | 1.56 (0.50-4.92) | 0.44 | 65% | 1.25 (0.84-1.86) | 0.28 | 0% |
| C combine | 1 | 0.51 (0.35-0.75) | 0.00 | -- | 0.46 (0.25-0.83) | 0.01 | -- | 0.55 (0.33-0.92) | 0.02 | -- |
C alone: cetuximab used as single agent; C combine: cetuximab used in combination with irinotecan or oxaliplatin based chemotherapy.
EGF A61G polymorphism (rs4444903) and toxicity
Pooled analysis result showed that EGF A61G polymorphism was not significantly associated with grades 2-3 cetuximab-related skin toxicity (RR = 1.25; 95% CI = 0.81-1.93) (Figure 2).
Figure 2.
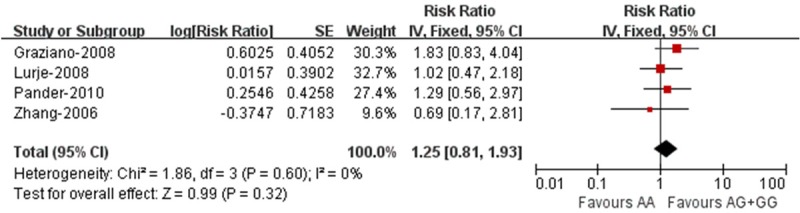
Association between EGF A61G polymorphism (rs4444903) and toxicity of cetuximab.
Publication bias
The publication bias was not considered to be significant (Figure 3).
Figure 3.
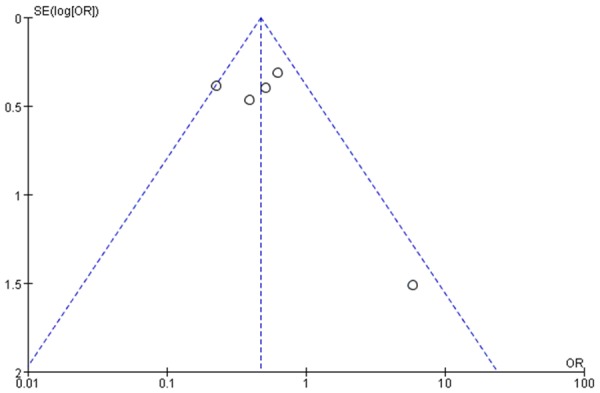
Funnel plot for publication bias.
Discussion
The present meta-analysis was performed to assess the potential value of EGF A61G polymorphism as predictive factors for cetuximab agents. Overall, we found substantial evidence that in mCRC this genetic variant is an appropriate marker for identifying a subgroup of patients who are more likely to have response rate with cetuximab.
EGF A61G polymorphism was widely investigated on the association with the risk to malignancies, including glioma [24,25], gastric cancer [26], hepatocellular carcinoma [27,28] as well as melanoma [29]. These evidence-based studies suggested that EGF 61G allele was likely to be associated with the risk of tumor susceptibility. On the other hand, GG genotype is related to less tumor recurrence in patients with esophageal cancers [30]. Every coin has two sides. EGF 61G allele played a paradoxical role of either increasing risk of cancer or improving prognosis of malignant disease. The mechanism remains to be defined.
In-vitro study has confirmed that cells carrying G allele produced significantly more EGF than cells with AA homozygotes [12]. Moreover, EGF could induce amphiregulin and epiregulin mRNA expression [31] both of which belong to epidermal growth factor family and are ligands to EGFR. Although epiregulin is known to bind less closely to EGFR than EGF, it can lead to prolonged state of receptor activation [32]. Khambata and his colleague [33] demonstrated that increased expression of epiregulin can promote tumor growth and survival, which may indicate the ability of cetuximab to block ligand-receptor interaction. All in all, patients with higher level expression of epiregulin and amphiregulin are more likely to have disease control with cetuximab, and the expression of epiregulin could be elevated by epidermal growth factor which synthesizes more in the cases harboring EGF 61G allele.
Our findings showed that patients carrying GG genotype showed a better response rate and prolonged PFS after being treated with cetuximab, which was in lined with those experiment results. Regarding of the association between variant and response rate to cetuximab, patients harboring GG homozygotes had a better clinical response rate than patients with AA homozygotes or AG heterozygotes. Concerning the association between EGF polymorphism and prognosis, as all studies only presented the HR and their 95% (CIs) of PFS or OS in M3 (AG+GG vs. AA). In our findings, GG carriers showed significantly prolonged PFS than both AA and AG carriers. Unfortunately, no better OS was found.
Almost all the studies included in this meta-analysis investigated the association between EGF A61G polymorphism and clinical outcomes independent of KRAS status, except for Pander’s research [22]. Pander and colleagues reported no significant correlation between this genetic variant and the efficacy of cetuximab for patients with KRAS wild-type metastatic colorectal cancer. As known, KRAS plays an important role in the RAS/MAPK pathway, and is involved in cell proliferation. The presence of activated KRAS mutations might circumvent cetuximab’s inhibitory activity.
Our statistical analysis showed that publication bias did not exist. However, there is no guarantee of the absence of bias, since the meta-analysis was based on a limited number of studies, which might reduce the chance of detecting publication bias to some extent [34,35]. Additionally, most of the studies had small sample sizes and variant previous treatments and concomitant medication. In addition, the criteria for assessing the response rate were different. Another limitation of current study is the confounding factors of KRAS status in patients with mCRC, which are complex and diverse. However, our review only included some of the confounding factors. Those unanalyzed factors that were inconsistent with our results might inevitably affect the presented results.
According to our meta-analysis, we would like to suggest further recommendations about relevant studies. To identify a biomarker in a more meaningful manner, additional trials should be conducted to value the influence of cetuximab on mCRC patients with mutated KRAS status. In addition, more high quality studies to evaluate the relationship between EGF polymorphism and cetuximab treated mCRC are warranted. Our meta-analysis has shown that the patients harboring GG homozygotes with mCRC received a better response rate after cetuximab treatment; however, this finding will determine whether the putative markers are predictive and/or prognostic and also whether they are associated with an improved clinical outcome measured by OS. Although there is still some space to improve, we consider that our study is powerful enough to show that EGF A61G polymorphism might predict improved clinical outcomes for mCRC patients treated with cetuximab.
In summary, this meta-analysis showed that mCRC patients harboring GG genotype of EGF A61G polymorphism are more likely to have a better clinical response with cetuximab treatment. The identified markers could be developed further to select patients for cetuximab therapy.
Acknowledgements
This work was supported by National Science Foundation of China (No. 81201888, 81372549 and No. 81172370) and Ministry of Education (IRT13101).
Disclosure of conflict of interest
None.
References
- 1.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 2.Kim ES, Khuri FR, Herbst RS. Epidermal growth factor receptor biology (IMC-C225) Curr Opin Oncol. 2001;13:506–513. doi: 10.1097/00001622-200111000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Benson AB 3rd, Bekaii-Saab T, Chan E, Chen YJ, Choti MA, Cooper HS, Engstrom PF, Enzinger PC, Fakih MG, Fenton MJ, Fuchs CS, Grem JL, Hunt S, Kamel A, Leong LA, Lin E, May KS, Mulcahy MF, Murphy K, Rohren E, Ryan DP, Saltz L, Sharma S, Shibata D, Skibber JM, Small W Jr, Sofocleous CT, Venook AP, Willett CG, Gregory KM, Freedman-Cass DA. Metastatic colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11:141–152. doi: 10.6004/jnccn.2013.0022. quiz 152. [DOI] [PubMed] [Google Scholar]
- 4.Wadler S. Targeted therapy in colorectal cancer. Clin Colorectal Cancer. 2007;6:357–361. doi: 10.3816/CCC.2007.n.004. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 6.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 8.Lenz HJ, Van Cutsem E, Khambata-Ford S, Mayer RJ, Gold P, Stella P, Mirtsching B, Cohn AL, Pippas AW, Azarnia N, Tsuchihashi Z, Mauro DJ, Rowinsky EK. Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan, oxaliplatin, and fluoropyrimidines. J. Clin. Oncol. 2006;24:4914–4921. doi: 10.1200/JCO.2006.06.7595. [DOI] [PubMed] [Google Scholar]
- 9.Saltz L. Systemic therapy for metastatic colorectal cancer. J Natl Compr Canc Netw. 2013;11:649–652. doi: 10.6004/jnccn.2013.0193. [DOI] [PubMed] [Google Scholar]
- 10.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 11.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 12.Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, Hutchinson PE, Osborne JE, Lear JT, Smith AG, Hutchinson IV. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet. 2002;359:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 13.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 14.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34:1319–1328. doi: 10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- 15.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28:123–137. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- 17.Song F, Khan KS, Dinnes J, Sutton AJ. Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Int J Epidemiol. 2002;31:88–95. doi: 10.1093/ije/31.1.88. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Gordon M, Press OA, Rhodes K, Vallbohmer D, Yang DY, Park D, Fazzone W, Schultheis A, Sherrod AE, Iqbal S, Groshen S, Lenz HJ. Cyclin D1 and epidermal growth factor polymorphisms associated with survival in patients with advanced colorectal cancer treated with Cetuximab. Pharmacogenet Genomics. 2006;16:475–483. doi: 10.1097/01.fpc.0000220562.67595.a5. [DOI] [PubMed] [Google Scholar]
- 19.Graziano F, Ruzzo A, Loupakis F, Canestrari E, Santini D, Catalano V, Bisonni R, Torresi U, Floriani I, Schiavon G, Andreoni F, Maltese P, Rulli E, Humar B, Falcone A, Giustini L, Tonini G, Fontana A, Masi G, Magnani M. Pharmacogenetic profiling for cetuximab plus irinotecan therapy in patients with refractory advanced colorectal cancer. J. Clin. Oncol. 2008;26:1427–1434. doi: 10.1200/JCO.2007.12.4602. [DOI] [PubMed] [Google Scholar]
- 20.Lurje G, Nagashima F, Zhang W, Yang D, Chang HM, Gordon MA, El-Khoueiry A, Husain H, Wilson PM, Ladner RD, Mauro DJ, Langer C, Rowinsky EK, Lenz HJ. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res. 2008;14:7884–7895. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
- 21.Garm Spindler KL, Pallisgaard N, Rasmussen AA, Lindebjerg J, Andersen RF, Cruger D, Jakobsen A. The importance of KRAS mutations and EGF61A>G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Ann Oncol. 2009;20:879–884. doi: 10.1093/annonc/mdn712. [DOI] [PubMed] [Google Scholar]
- 22.Pander J, Gelderblom H, Antonini NF, Tol J, van Krieken JH, van der Straaten T, Punt CJ, Guchelaar HJ. Correlation of FCGR3A and EGFR germline polymorphisms with the efficacy of cetuximab in KRAS wild-type metastatic colorectal cancer. Eur J Cancer. 2010;46:1829–1834. doi: 10.1016/j.ejca.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Hu-Lieskovan S, Vallbohmer D, Zhang W, Yang D, Pohl A, Labonte MJ, Grimminger PP, Holscher AH, Semrau R, Arnold D, Dellas K, Debucquoy A, Haustermans K, Machiels JP, Sempoux C, Rodel C, Bracko M, Velenik V, Lenz HJ. EGF61 polymorphism predicts complete pathologic response to cetuximab-based chemoradiation independent of KRAS status in locally advanced rectal cancer patients. Clin Cancer Res. 2011;17:5161–5169. doi: 10.1158/1078-0432.CCR-10-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan D, Xu J, Li Y, Lai R. Association between +61G polymorphism of the EGF gene and glioma risk in different ethnicities: a meta-analysis. Tohoku J Exp Med. 2010;222:229–235. doi: 10.1620/tjem.222.229. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Xi L, Zeng J, Yao Q. A functional +61G/A polymorphism in epidermal growth factor is associated with glioma risk among Asians. PLoS One. 2012;7:e41470. doi: 10.1371/journal.pone.0041470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piao Y, Liu Z, Ding Z, Xu L, Guo F, Sun Q, Xie X. EGF+61A>G polymorphism and gastrointestinal cancer risk: a HuGE review and meta-analysis. Gene. 2013;519:26–33. doi: 10.1016/j.gene.2013.01.057. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Wu Q, Shi Y, Nie Y, Wu K, Fan D. Epidermal growth factor 61A>G polymorphism is associated with risk of hepatocellular carcinoma: a meta-analysis. Genet Test Mol Biomarkers. 2012;16:1086–1091. doi: 10.1089/gtmb.2012.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong JH, You XM, Gong WF, Ma L, Zhang Y, Mo QG, Wu LC, Xiao J, Li LQ. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;7:e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu D, Wu Y, Zhang X, Cong P, Lv X. Lack of association between EGF+61A>G polymorphism and melanoma susceptibility in Caucasians: a HuGE review and meta-analysis. Gene. 2013;515:359–366. doi: 10.1016/j.gene.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Lurje G, Leers JM, Pohl A, Oezcelik A, Zhang W, Ayazi S, Winder T, Ning Y, Yang D, Klipfel NE, Chandrasoma P, Hagen JA, DeMeester SR, DeMeester TR, Lenz HJ. Genetic variations in angiogenesis pathway genes predict tumor recurrence in localized adenocarcinoma of the esophagus. Ann Surg. 2010;251:857–864. doi: 10.1097/SLA.0b013e3181c97fcf. [DOI] [PubMed] [Google Scholar]
- 31.Inatomi O, Andoh A, Yagi Y, Bamba S, Tsujikawa T, Fujiyama Y. Regulation of amphiregulin and epiregulin expression in human colonic subepithelial myofibroblasts. Int J Mol Med. 2006;18:497–503. [PubMed] [Google Scholar]
- 32.Shelly M, Pinkas-Kramarski R, Guarino BC, Waterman H, Wang LM, Lyass L, Alimandi M, Kuo A, Bacus SS, Pierce JH, Andrews GC, Yarden Y. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J Biol Chem. 1998;273:10496–10505. doi: 10.1074/jbc.273.17.10496. [DOI] [PubMed] [Google Scholar]
- 33.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA 3rd, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J. Clin. Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]


