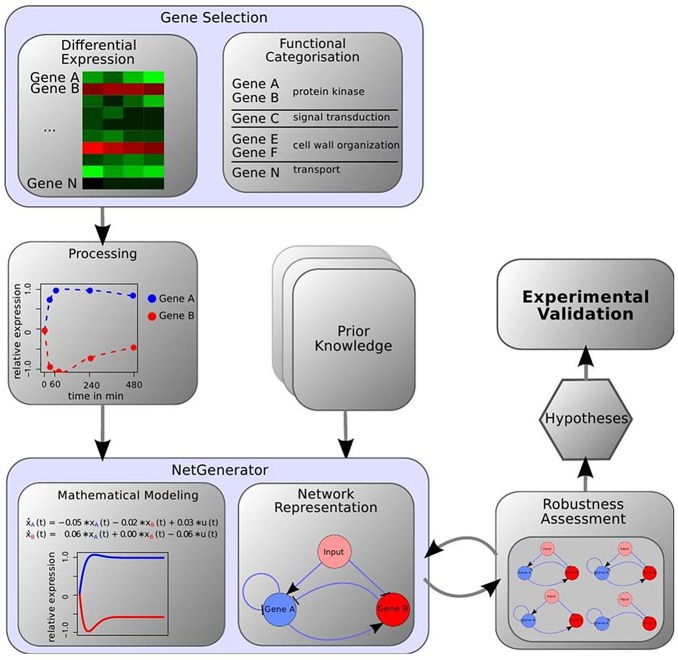
Fig 1. Depiction of the workflow.
Genes were selected based on their expression steady-state levels and their assigned function. RNA-Seq data and prior-knowledge were used as inputs for the NetGenerator. Using a mathematical modeling, a network was predicted, which was then evaluated and tested for robustness. A final model was selected, which led to new hypotheses that were experimentally validated.