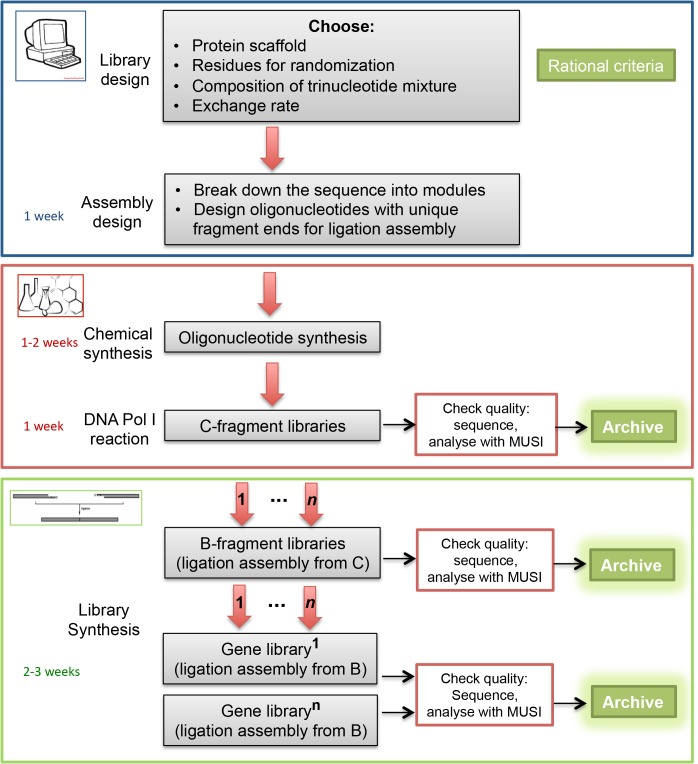
Fig 8. Summary workflow diagram of the method for combinatorial gene synthesis.
Library design: the method imposes no restrictions to the number of residues selected for mutagenesis or to their location. The composition of the trinucleotide mixture and the exchange rate can be freely chosen without any limitations. Contact the oligonucleotide synthesis company of your choice well in advance and coordinate the purchase of trimer phosphoramidites. Assembly design: The fractionation of the gene sequence into modules depends on the locations of the residues for randomization. Divide the gene sequence into modules with a length between 40 bp and 90 bp, containing the mutagenized codons. The fragment borders of the modules should create unique overhands for ligation assembly after restriction digestion (see Table 1 and main text for details). Use PCR for longer stretches of wild type gene sequence. Chemical synthesis: Invest efforts and resources into high quality oligonucleotide synthesis as well as cloning and analysis of the C-libraries. The chemically synthesized diversity is stored in them and they are the starting point for all future gene libraries. Clone a wild type sequence, corresponding to each mutagenized module. Library synthesis: Generate B-fragment libraries by ligation from C-fragment libraries. Different B-fragment libraries (1, …, n) can be obtained in parallel by exchanging or mixing mutagenized modules with the corresponding wild type modules. Archive the B-libraries. Generate gene libraries by ligation assembly of B-libraries. Use the combinatorics approach for generation of multiple unique gene libraries.