Summary
The synaptonemal complex (SC) is a conserved protein complex that stabilizes interactions along homologous chromosomes (homologs) during meiosis. The SC regulates genetic exchanges between homologs, thereby enabling reductional division and the production of haploid gametes. Here we directly observe SC assembly (synapsis) by optimizing methods for long-term fluorescence recording in C. elegans. We report that synapsis initiates independently on each chromosome pair at or near Pairing Centers – specialized regions required for homolog associations. Once initiated, the SC extends rapidly and mostly irreversibly to chromosome ends. Quantitation of SC initiation frequencies and extension rates reveals that initiation is a rate-limiting step in homolog interactions. Eliminating the dynein-driven chromosome movements that accompany synapsis severely retards SC extension, revealing a new role for these conserved motions. This work provides the first opportunity to directly observe and quantify key aspects of meiotic chromosome interactions, and will enable future in vivo analysis of germline processes.
Introduction
Meiosis is the specialized cell division program that produces haploid gametes from diploid precursors, and is therefore essential for sexual reproduction. A unique aspect of meiosis is that homologous parental chromosomes (homologs) are partitioned into different daughter cells. This reductional segregation requires physical connections to be established between homologs during the protracted meiotic prophase that precedes the meiotic divisions. Pairing between homologs enables the formation of linkages known as chiasmata, which are products of crossover recombination. However, recombination can lead to chromosome rearrangements if it occurs ectopically, i.e., between homologous sequences that are not syntenic. Alignment of homologs along their entire lengths may help to ensure that exchanges occur only between homologous sequences on appropriate partner chromosomes.
Chromosomes achieve intimate side-by-side alignment in a stepwise fashion. During early meiotic prophase, special sites on each chromosome establish connections through the nuclear envelope to the microtubule cytoskeleton (or actin cables, in budding yeast) and initiate movements along the nuclear periphery through the action of dynein and/or other motors (Hiraoka and Dernburg, 2009; Rog and Dernburg, 2013). In most species this is a meiotic function of telomeres, but in C. elegans this role has been acquired by a region near one end of each chromosome known as a Pairing Center (PC) (MacQueen et al., 2005; Sato et al., 2009). The resulting large-scale chromosome motions facilitate homolog pairing, although the mechanisms by which homology is recognized remain unclear.
Assembly of the SC (also known as synapsis) is required to achieve stable, length-wise homolog alignment (MacQueen et al., 2002; Page and Hawley, 2001; Sym et al., 1993). In many organisms, initiation of synapsis depends on recombination intermediates, which are hypothesized to trigger SC assembly by mediating long-lived local DNA base-pairing interactions (Page and Hawley, 2004). However, recombination-independent mechanisms also bring homologs into proximity in early prophase. For example, in mouse spermatocytes homologous chromosome territories become juxtaposed even in the absence of breaks or synapsis (Ishiguro et al., 2014). In the nematode C. elegans and the dipteran Drosophila melanogaster, recombination-independent pairing mechanisms have acquired greater robustness, to the extent that homolog pairing and synapsis occur normally in the absence of recombination (Dernburg et al., 1998; McKim et al., 1998). Whether local pairing is mediated by recombination or other mechanisms, how chromosomes progress from initial, localized associations to complete synapsis and lengthwise alignment remains poorly understood.
A major challenge in understanding the mechanisms that contribute to homolog interactions is that pairing and synapsis occur over many hours or days, commonly within internal reproductive organs. Another experimental obstacle is that pairing and synapsis occur between intact chromosomes as they interact with the nuclear envelope, precluding straightforward reconstitution of these processes in vitro or in cell-free systems. Due to these challenges, our current understanding is largely based on cytological analysis of fixed meiocytes, combined with molecular genetics. To gain insight into the dynamics of homologous chromosome interactions, we developed quantitative time-lapse imaging methods that enabled us to interrogate synapsis in living animals.
Results and Discussion
C. elegans offers many advantages for analysis of germline processes, including powerful molecular genetics, organization of nuclei within a syncytial gonad in a spatio-temporal gradient, and a simple karyotype (2N=12). Adult hermaphrodites are transparent and continuously undergo oogenesis, enabling in vivo imaging of meiotic nuclei (Baudrimont et al., 2010; Wynne et al., 2012). Standard methods for immobilizing living worms for imaging use paralytic drugs and/or friction, but we and others have found that these approaches result in an arrest of meiotic nuclei (Kim et al., 2013; Wynne et al., 2012). This arrest is evident by cessation of the dynein-driven chromosome motions that normally occur during early prophase (Wynne et al., 2012). We found that even in the absence of paralytic drugs or exposure to fluorescence illumination, worm immobilization led to arrest across the gonad, typically within 15–30 minutes (Kim et al., 2013) (n=46; Figure S1A and Movie 1). While this limited temporal window has been sufficient to characterize some aspects of prophase chromosome dynamics (Baudrimont et al., 2010; Wynne et al., 2012), the process of synapsis occurs over several hours and was therefore inaccessible to in vivo imaging.
We sought to mitigate the experimentally-induced arrest to enable long-term imaging of germline processes. We speculated that immobilization might induce this arrest because it impairs eating. Nutrient withdrawal rapidly triggers behavioral and metabolic changes in C. elegans through serotonergic signaling (Chase and Koelle, 2007; Flavell et al., 2013), so we tested whether the arrest could be suppressed by serotonin. Consistent with this idea, we found that immobilization in the presence of serotonin allowed chromosome motion to continue unabated for much longer (42.8% of animals after 1 hour [n=320]; Figure S1B–C and Movie 2). Throughout this work, we visualized immobilized worms supplemented with serotonin using a spinning-disc confocal microscope, which enabled long-term high-resolution multicolor 3D imaging without apparent phototoxicity (see below).
We also developed fluorescent reporters suitable for long-term imaging of synapsis. The C. elegans SC is comprised of at least four proteins that assemble at the interface between paired axes: SYP-1, -2, -3 and -4. Each requires the other three to associate with meiotic chromosomes (Colaiacovo et al., 2003; MacQueen et al., 2002; Smolikov et al., 2007; Smolikov et al., 2009). We engineered worms carrying a single-copy syp-3 transgene tagged with the Emerald variant of green fluorescent protein (hereafter EmGFP-SYP-3), which was validated as a reporter by a variety of assays for meiotic progression and chromosome segregation (Figure S2). Most experiments described below were carried out using hermaphrodites expressing EmGFP-SYP-3 and lacking wild-type SYP-3, which maximized the fluorescence associated with the SC, but since we detected slight meiotic perturbation in this strain we validated our key conclusions in strains exhibiting no meiotic defects (see below).
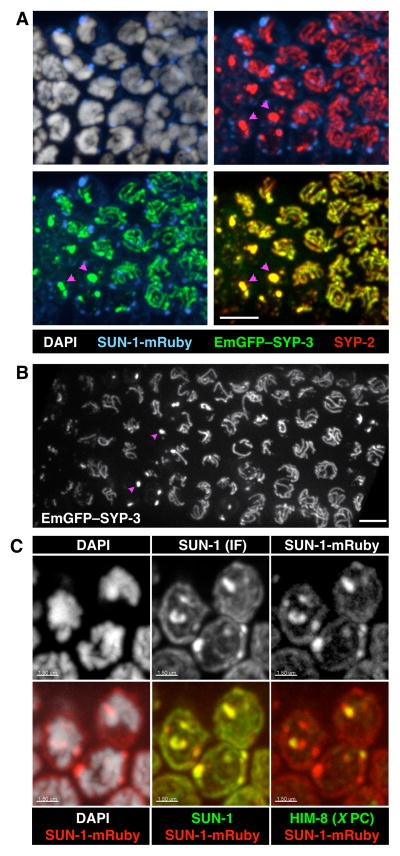
Upon entering meiosis, nuclei traverse the “transition zone” region of the gonad. This zone corresponds to the classically defined stages of leptonema and zygonema, during which homologs pair and synapse. Transition zone nuclei are marked by asymmetrical distribution of chromosomes within the nucleus and the aggregation of the nuclear envelope proteins SUN-1 and ZYG-12 as they associate with PCs. This SUN/KASH pair connects chromosomes to the microtubule cytoskeleton (Hiraoka and Dernburg, 2009; Sato et al., 2009). In fixed gonads, EmGFP-SYP-3 localization mirrored that of endogenous SYP-3: in the earliest stages of meiosis, EmGFP-SYP-3 was detected in small nuclear aggregates, and as nuclei traversed the transition zone it formed multiple filaments, each at the interface between a pair of homologs (Figure 1A–B and Figure S3A, and Movie 3).
Figure 1. Fluorescent reporters for the synaptonemal complex.
(A) Maximum intensity projections of 3D images of the transition zone region of a fixed gonad from an EmGFP-SYP-3; syp-3; SUN-1-mRuby hermaphrodite, stained with antibodies against SYP-2 and DAPI. EmGFP-SYP-3 and SUN-1-mRuby were visualized by their intrinsic fluorescence. Meiosis progresses from left to right in these images. Each image shows two superimposed channels. Pink arrowheads point to examples of SC aggregates (polycomplexes). Scale bar = 5 μm. See also Figures S1 & S2.
(B) Projection of the transition zone region from a live EmGFP-SYP-3; syp-3 worm. Meiotic progression is from left to right. Pink arrowheads mark examples of polycomplexes. Scale bar = 5 μm. A 3D rendering of the same image stack is shown in Movie 3. See Figure S3A for an image of an entire gonad.
(C) Maximum intensity projections of transition zone nuclei from a SUN-1-mRuby adult hermaphrodite stained with antibodies against SUN-1 and HIM-8, and DAPI. SUN-1-mRuby was visualized by its intrinsic fluorescence. SUN-1 immunofluorescence colocalizes with SUN-1-mRuby. HIM-8, marking the X chromosome PCs, colocalizes with one of the SUN-1 patches. Scale bars = 1.5 μm. See Figure S3B for an image of an entire gonad.
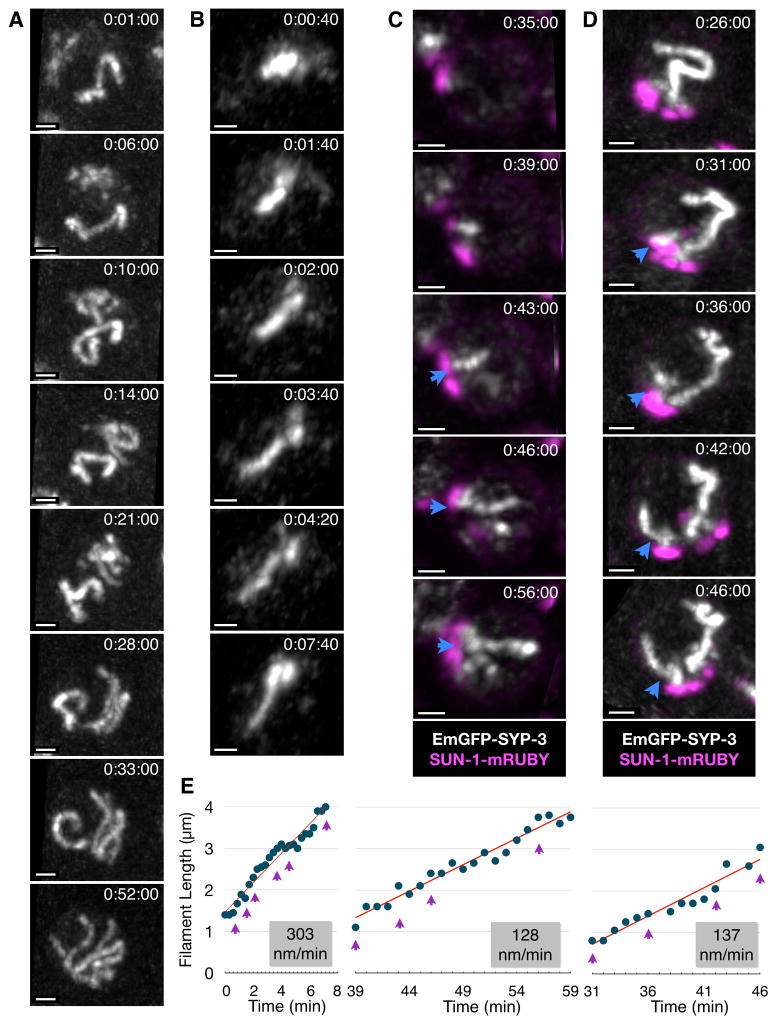
These fluorescently tagged proteins, together with our long-term immobilization technique, allowed us to directly observe and analyze the assembly of SCs in vivo. Within transition zone nuclei, we observed many examples of individual SC filaments that rapidly extended after they were first detected. These SCs usually appeared one at a time within a nucleus, but occasionally two or three filaments appeared in close succession (Figure 2A–B and Movies 4&5). We performed manual tracing of filaments within 3D image stacks to determine their contour lengths (Figure S1D). It was evident from inspection of these data that each filament underwent growth at a fairly constant rate, eventually reaching a maximum length 4–6μm, similar to the length of complete SCs measured in fixed samples (Figures 2B–D & 3A). To determine the rates of SC extension for individual filaments, their lengths as a function of time were analyzed by segmented linear regression. The median rate was 150nm/min (mean=170; SD=70; range 80–300), corresponding to complete synapsis of a chromosome pair in 25–40 mins (range 13–75 mins). We obtained comparable rates from animals that expressed wild-type SYP-3 in addition to the tagged protein, indicating that absence of untagged SYP-3 does not alter the kinetics of SC extension (Figure 3B–C). SC elongation rates were not markedly affected by the time elapsed since the beginning of image acquisition (Figure 3A) or the image acquisition rate (Figure 3B), indicating that SC assembly is not highly photosensitive under our conditions. Neither nascent nor fully formed SC filaments underwent significant shortening beyond small fluctuations, likely reflecting measurement errors (e.g., Figure 3A), indicating that large-scale de-synapsis events are rare and that synapsis is likely irreversible, at least once a filament has reached a threshold length that permits detection.
Figure 2. In vivo imaging of SC assembly.
(A) Maximum intensity projections showing a single nucleus at selected timepoints from a series of 3D images acquired at 1-minute intervals. One complete SC is present at the beginning of image acquisition, and new filaments appear between timepoints 0:06:00 and 0:33:00. The full recording is shown in Movie 4. Time elapsed since the beginning of image acquisition is reported as h:mm:ss. Scale bar = 1 μm.
(B) Projections from selected timepoints showing a single nucleus from a 3D image series acquired at 20-second intervals; the full recording is shown in Movie 5. Scale bar = 1 μm. Bottom, a plot showing contour length of the SC filament as a function of time. The slope of the elongating portion of the plot based on segmented linear regression is shown in red. Purple arrowheads indicate values for the timepoints shown above.
(C–D) Partial Z projections from selected timepoints showing nuclei from EmGFP-SYP-3; syp-3; SUN-1-mRuby hermaphrodites. 3D images were acquired at 1-minute intervals. In each series, a nascent SC filament is marked with a blue arrowhead. Scale bars = 1 μm. Filament lengths over time are plotted below. The slope of the elongating portion of the plot is shown in red. Purple arrowheads indicate the time points represented in the images above. The corresponding full time courses are included as Movie 6(C) and 7(D).
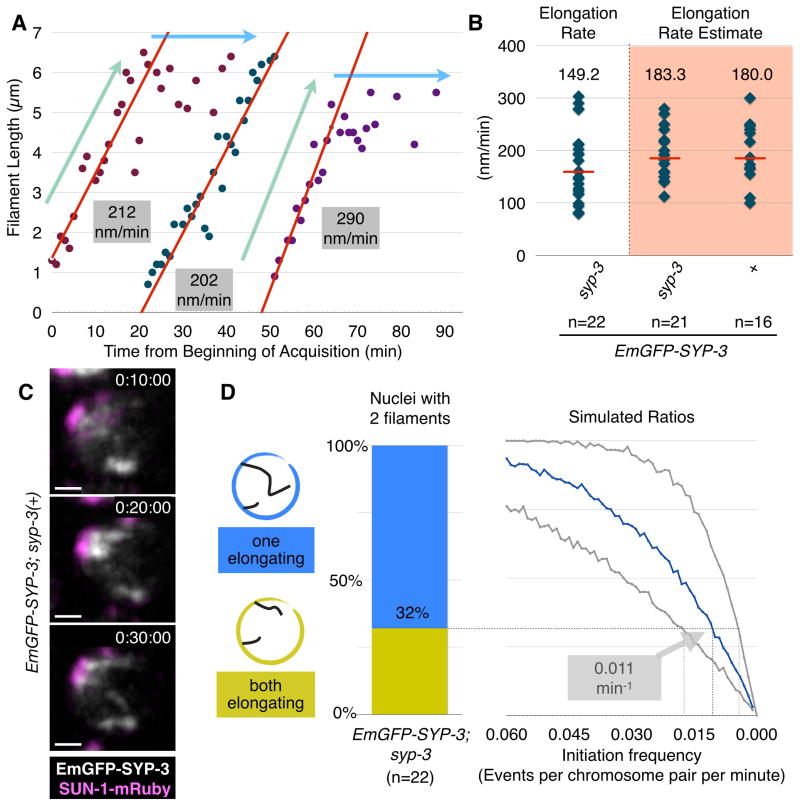
Figure 3. Quantification of SC initiation and extension.
(A) Plots of elongating SC filaments in selected nuclei. Data for each SC is shown in a different color. The ‘elongating’ and ‘constant’ portions of the plots are designated with light blue and green arrows, respectively. The portion of each plot from which the elongation rate was determined is shown as a red line. Notably, new filaments appear and elongate at similar rates even after more than 50 mins of imaging, indicating that SC assembly is not markedly affected by our imaging conditions.
(B) Plots showing the distribution of SC elongation rates based on many individual examples from the indicated genotypes. Median values are indicated by red bars and their numerical values are reported above the distribution. The shaded distributions reflect estimated rates based on analysis at lower temporal resolution. Note that under the slower acquisition conditions, animals were subjected to 10–20-fold less illumination light, yet the elongation rate was equivalent to that measured in data sets with higher temporal resolution, indicating that SC assembly is not highly photosensitive under our conditions.
(C) An example of synapsis progression in an EmGFP-SYP-3; SUN-1-mRuby hermaphrodite imaged at low temporal resolution (every 10 minutes). Two nascent filaments appear between timepoints 0:10:00 and 0:30:00. Scale bar = 1 μm.
(D) Estimation of initiation rates based on analysis of fixed nuclei. We identified nuclei with two visible SC filaments in high-resolution 3D images of fixed gonads stained with antibodies against SC proteins, and classified each filament as either ‘partial’ (<3.5μm) or ‘complete’ (≥3.5μm). Left, the ratio of nuclei with two incomplete (elongating) filaments versus nuclei with one complete filament and one elongating filament. Right, expected ratios derived from simulations with varying initiation frequencies and elongation rates; the x-axis indicates the simulated initiation frequency, while the different curves correspond to different elongation rates: the blue curve reflects the median measured rate (149nm/min), while the gray curves correspond to the minimum and maximum observed elongation rates (80 and 300 nm/min, respectively). Our measured rate of SC elongation and the ratio between nuclei harboring one versus two elongating filaments yield an initiation frequency of 0.011 per chromosome per minute (range: 0.005–0.019). See also Figure S4.
Previous studies have demonstrated that SC assembly in C. elegans normally depends on PCs, and that homology within the PC regions is sufficient for otherwise nonhomologous chromosomes to synapse (Hayashi et al., 2010; MacQueen et al., 2005; Penkner et al., 2007; Sato et al., 2009). These and other observations have suggested that PCs act as sites of synapsis initiation, although this idea has not been directly tested. We marked PCs by expressing the nuclear envelope protein SUN-1 fused at its C-terminus to the monomeric red fluorescent protein mRuby (hereafter SUN-1-mRuby), and crossed this marker into our SC-labeled strain (Figure 1C and Figure S3B). In nuclei from animals expressing both fluorescent reporters, nascent SC filaments were always apposed to a patch of SUN-1 (n=28; Figure 2C–D and Movies 6&7), providing direct evidence of initiation at or near PCs. Together with the quantitative analysis above, these observations demonstrate that synapsis of each homolog pair initiates only once, and that PCs are the primary sites for initiation. Once nucleated, the SC extends along the chromosome at a constant rate.
Next we wished to investigate the dynamics of synapsis throughout the nucleus, and to quantify the rates of synapsis elongation relative to initiation and to pairing at PCs. Our ability to track and measure SC filaments was facilitated by the sparseness of SCs during early stages of synapsis (e.g., Figures 1&2). This observation indicates that initiation of SC assembly on different chromosome pairs is staggered and infrequent relative to the rate of extension. We used two independent methods to determine the frequency of synapsis initiation, which yielded highly concordant values. First, we measured the time that elapsed between the first and second initiation events in nuclei where these events were clearly observed in our time-lapse recordings (e.g., Figure 3C), and found that the average interval was 22 minutes (SD=15; median=21; n=12). Since there are 5 unsynapsed homolog pairs at this stage, this corresponds to an initiation frequency of 0.009 events per chromosome pair per minute if we assume that initiation occurs independently for each pair. Second, among nuclei in which 2 SC filaments were observed in high resolution fixed images, we determined how many contained two elongating filaments versus one elongating and one complete filament (Figure 3D). This ratio, combined with our measured SC extension rate obtained by in vivo imaging, yielded a very similar initiation rate of 0.011 events per pair per minute. Importantly, pairwise association between homologous PCs was not rate-limiting for synapsis, since homologously paired PCs lacking SCs were often observed in fixed images (Figure S4A–B).
We found that the six chromosome pairs do not synapse in a stereotyped order (Figure S4A–B), consistent with prior analysis (Nabeshima et al., 2011). Additional simulations and observations of fixed nuclei validated our assumption that the probability of initiation per unit time is proportional to the number of unsynapsed pairs, and therefore that the likelihood that any given pair will initiate synapsis is largely independent of the number of pairs that have already synapsed (Figure S4C). These observations, together with our measured synapsis initiation frequency and synapsis elongation rate, allowed us to simulate the process of synapsis for whole nuclei. We found that a median time of 3.4–4.1 h is required for all six homolog pairs to initiate synapsis, and that 3.9–4.6 h are necessary to complete synapsis (allowing 30 minutes for the last pair to fully synapse). This number is in good agreement with the estimated 5 hours spent in the transition zone, which nuclei exit once all chromosomes achieve synapsis (Crittenden et al., 2006; Fox et al., 2011). Together, our observations are consistent with the idea that initiation of synapsis between each pair of homologs is a stochastic process that occurs independently of the synapsis status of other chromosomes.
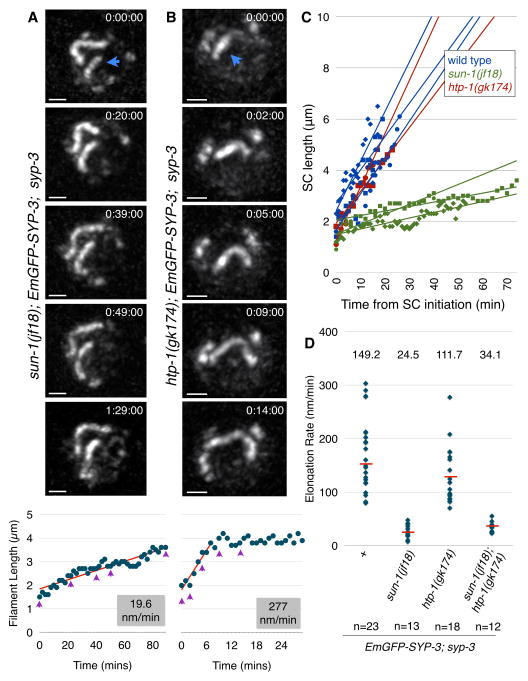
Prior analysis of fixed samples has shown that stable pairing along the length of homologs requires synapsis, while pairing at the PC region is synapsis-independent (MacQueen et al., 2002). We wondered whether elongation of the SC simply stabilizes interactions between axes as they bump into each other, or if instead SC assembly generates forces that actively draw paired chromosomes into full alignment. To test this we explored whether PC-led chromosome motion, which elongates chromosomes and contributes to alignment (Nabeshima et al., 2011), might affect the formation of the SC. We have previously shown that the activity of dynein generates long-range, processive chromosome motions that enhance the rate of homolog pairing at PCs (Sato et al., 2009; Wynne et al., 2012). Because dynein-driven forces are normally necessary for synapsis initiation, we tested the role of these motions in SC extension by comparing two mutants: sun-1(jf18) and htp-1(gk174). In both of these mutants synapsis occurs in the absence of dynein, but SC forms primarily between nonhomologous chromosomes (Baudrimont et al., 2010; Penkner et al., 2007; Sato et al., 2009; Wynne et al., 2012). However, the two mutations have very different effects on dynein-driven chromosome motion, which is abrogated in sun-1(jf18) mutants but only subtly reduced in htp-1(gk174) (Baudrimont et al., 2010; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). We found that SC extension was severely retarded in sun-1(jf18) hermaphrodites but occurred at nearly the wild-type rate in htp-1(gk174) mutants (sun-1: median=25nm/min; mean=26; SD=12; range 8–48; Figure 4A,C&D and Movie 8; htp-1: median=110nm/min; mean=130; SD=55; range 70–280; Figure 4B–D and Movie 9). The rate of SC extension in sun-1(jf18); htp-1(gk174) double-mutants, which lack active chromosome movement, was similar to sun-1(jf18) single mutants (median=34nm/min; mean=34; SD=10; range 23–55; Figure 4D). The slightly reduced rate in htp-1 compared to wild-type may reflect slower and/or less frequent motion in htp-1(gk174) animals (Baudrimont et al., 2010), or a minor contribution of homology to the efficiency of synapsis extension. Interestingly, in all genotypes examined the rate of elongation was quite consistent along the length of chromosomes. Taken together, these results suggest that chromosome movement promotes SC extension. Because SUN-1 is embedded in the nuclear membrane, it is unlikely to directly contribute to extension of the SC along paired chromosomes, but we cannot rule out the possibility that SC elongation is inhibited in sun-1(jf18) mutants for reasons other than reduction of chromosome motion.
Figure 4. Dynein-driven chromosome motion promotes SC elongation.
(A–B) Partial Z-projections of selected timepoints of nuclei imaged every 1 min. The nascent filament is marked with a blue arrowhead. Scale bars = 1 μm. Filament lengths over time are plotted below. The slope of the elongating portion of the plot is shown in red. Purple arrowheads indicate the timepoints shown above.
(A) A nucleus from an EmGFP-SYP-3; syp-3; sun-1(jf18) hermaphrodite. Note that the two synapsed chromosomes barely move relative to each other, reflecting the absence of dynein-driven chromosome motions (the full recording is shown in Movie 8).
(B) A nucleus from an EmGFP-SYP-3; syp-3; htp-1(gk174) hermaphrodite (the full recording is shown in Movie 9).
(C) Representative plots of elongating filaments from the indicated genotypes. Times are relative to the initiation of SC assembly.
(D) Distributions of SC elongation rates measured from the indicated genotypes. The median is indicated by a red bar and its numerical value is reported above the distribution.
We speculate that PC-led chromosome motion may facilitate SC extension by creating hydrodynamic forces that act along the chromosomes to promote an elongated, roughly parallel configuration, and/or to reduce the distance or increase collision frequencies between axes. SC assembly may mediate alignment by sequential stabilization of local collisions, thereby acting as a ratchet (Movie 10). In the absence of chromosome motions, as in the sun-1(jf18) mutant, SC assembly may still act as a Brownian ratchet, relying primarily on thermal fluctuations to bring unsynapsed axes adjacent to the growing SC into proximity. Dragging chromosomes through the nucleoplasm may also promote SC assembly by facilitating the removal of intervening obstacles, such as other chromosomes. However, the absence of long pauses during SC elongation even in sun-1(jf18) mutants suggests that such obstructions do not typically persist for longer than a few minutes.
Our refinement of imaging methods for C. elegans has enabled us to further harness the potential of this model system, and to directly observe and quantify the dynamic process of synapsis. We find that synapsis initiates at or near PCs and then proceeds to ‘zip’ together entire chromosomes at a rapid and constant rate. Furthermore, we show that dynein-driven chromosome motions greatly accelerate the rates of SC assembly and chromosome alignment, adding to our understanding of the functions of these unique meiotic chromosome dynamics. Our findings directly demonstrate that synapsis is largely irreversible and is a rate-limiting step in the homologous chromosome interactions necessary for faithful execution of meiosis. The tools we have developed also open the door to investigate a variety of cellular and subcellular dynamics in this experimental system.
Experimental Procedures
All strains were maintained using standard methods (Brenner, 1974). Immunofluorescence was performed as previously described (Phillips et al., 2009). Single-copy transgenes were generated by MosSCI (Frokjaer-Jensen et al., 2012; Frokjaer-Jensen et al., 2008). For live imaging, adult hermaphrodites were immobilized on freshly prepared 7.5% agarose pads overlayed with 100nm polystyrene beads (Kim et al., 2013) and serotonin at a final concentration of 25mM. Worms were overlaid with coverslips that were sealed to the slide, and imaged immediately afterwards. Images were acquired using a spinning-disc confocal microscope at ambient temperature, using a 63× 1.4NA oil objective at 0.5μm z-spacing. Time-lapse series were segmented, tracked and aligned based on overall fluorescence of EmGFP-SYP-3. Filaments in informative nuclei were traced by hand from 3D reconstructions. SC length plots were subjected to segmented linear regression. Simulations were performed by considering the synapsis state of 6 homolog pairs using custom code. Full details are available in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Y. Kim for purification of the SYP-2 antibody, T. Barros for help with producing Movie 10, D.J. Wynne for innovations in live-imaging technology and S. Nakielny, Y. Mazor, J.P. Cooper, R. Heald and members of our laboratory for discussion and critical reading of the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was supported by a European Molecular Biology Organization Long-Term Fellowship (ALTF 564-2010) to OR, and support to AFD from the National Institutes of Health (R01 GM065591), and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, et al. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001219. doi: 10.1371/journal.pgen.1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase DL, Koelle MR. Biogenic amine neurotransmitters in C. elegans. WormBook; 2007. pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Couteau F, Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, Bargmann CI. Serotonin and the Neuropeptide PDF Initiate and Extend Opposing Behavioral States in C. elegans. Cell. 2013 doi: 10.1016/j.cell.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PM, Vought VE, Hanazawa M, Lee MH, Maine EM, Schedl T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011;138:2223–2234. doi: 10.1242/dev.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Mlynarczyk-Evans S, Villeneuve AM. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics. 2010;186:45–58. doi: 10.1534/genetics.110.115501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Shibuya H, Hernandez-Hernandez A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, et al. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One. 2013;8:e53419. doi: 10.1371/journal.pone.0053419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Mlynarczyk-Evans S, Villeneuve AM. Chromosome painting reveals asynaptic full alignment of homologs and HIM-8-dependent remodeling of X chromosome territories during Caenorhabditis elegans meiosis. PLoS Genet. 2011;7:e1002231. doi: 10.1371/journal.pgen.1002231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Phillips CM, McDonald KL, Dernburg AF. Cytological analysis of meiosis in Caenorhabditis elegans. Methods Mol Biol. 2009;558:171–195. doi: 10.1007/978-1-60761-103-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Dernburg AF. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr Opin Cell Biol. 2013;25:349–356. doi: 10.1016/j.ceb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Eizinger A, Schild-Prufert K, Hurlburt A, McDonald K, Engebrecht J, Villeneuve AM, Colaiacovo MP. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 2007;176:2015–2025. doi: 10.1534/genetics.107.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Schild-Prufert K, Colaiacovo MP. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 2009;5:e1000669. doi: 10.1371/journal.pgen.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol. 2012;196:47–64. doi: 10.1083/jcb.201106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.