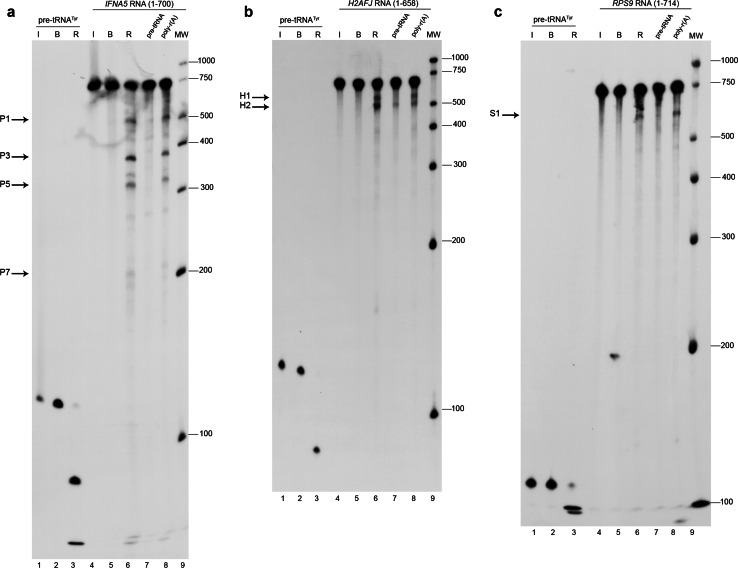
Fig. 2.
Specific cleavage of IFNA5, H2AFJ and RPS9 RNAs by human RNase P. All transcripts were internally radiolabelled during in vitro transcription. The RNA substrate for lanes 1–3 for all panels is the pre-tRNATyr of E. coli. Lanes 4–9 correspond to: IFNA5 RNA (1–700) for a, H2AFJ RNA (1–658) for b and RPS9 RNA (1–714) for c. In all autoradiograms: lanes 1 and 4 correspond to RNAs incubated on ice (I); lanes 2 and 5 in the presence of reaction buffer (B); lanes 3 and 6 reaction with human RNase P (R). Lane 7 human RNase P reaction incubated with a tenfold molar excess of cold pre-tRNATyr. Lane 8 incubated with 400 pg of a non-specific competitor poly-r(A). This corresponds to the amount of RNA by weight used in the tenfold pre-tRNATyr treatment in lane 7. Lane 9 molecular weight markers. The main digestion products are indicated by arrows on the left of each panel. The autoradiograms correspond to denaturing polyacrylamide gels at 4 %